Using topology to analyze the shape of barley


Preprint [QR below]
doi.org/10.1101/2021.03.27.437348


1 Computational Math, Science & Engineering, Michigan State University
2 Horticulture, Michigan State University
3 Mathematics and Computer Science, TU Eindhoven
4 Integrative Plant Science, Cornell University
5 Botany and Plant Sciences, University of California, Riverside


| Shape descriptors | # descr | F1 Score |
|---|---|---|
| Traditional | 11 | 0.55 ± 0.019 |
| Topological (ECT + UMAP) | 12 | 0.74 ± 0.016 |
| Combined (Trad + Topo) | 23 | 0.86 ± 0.010 |
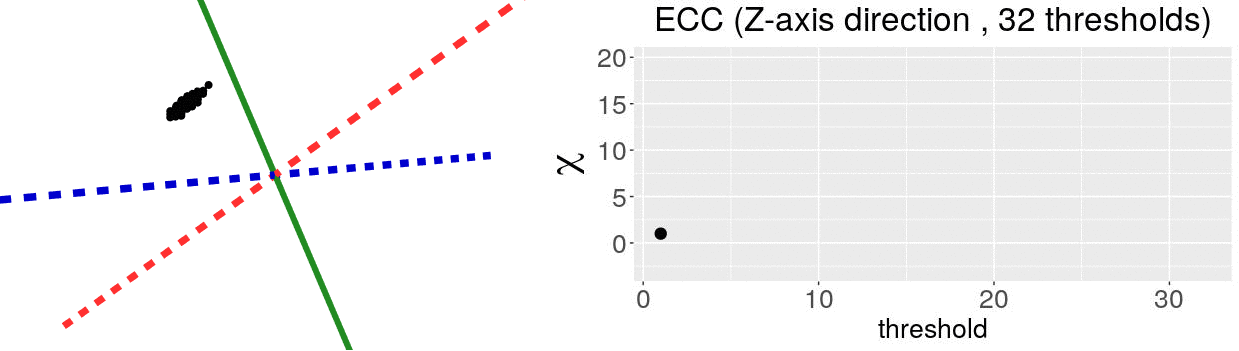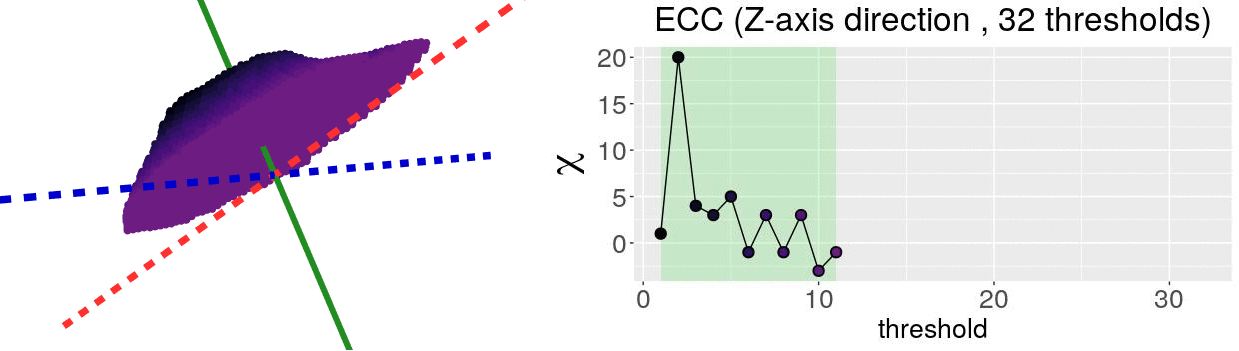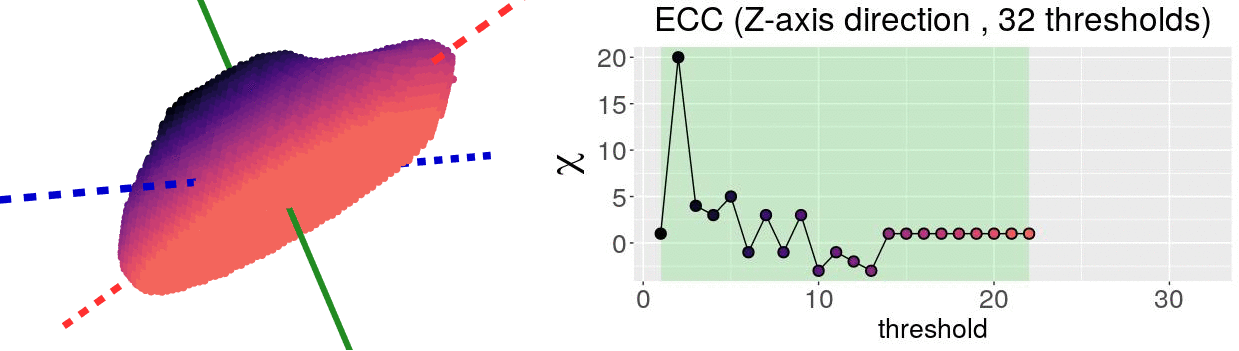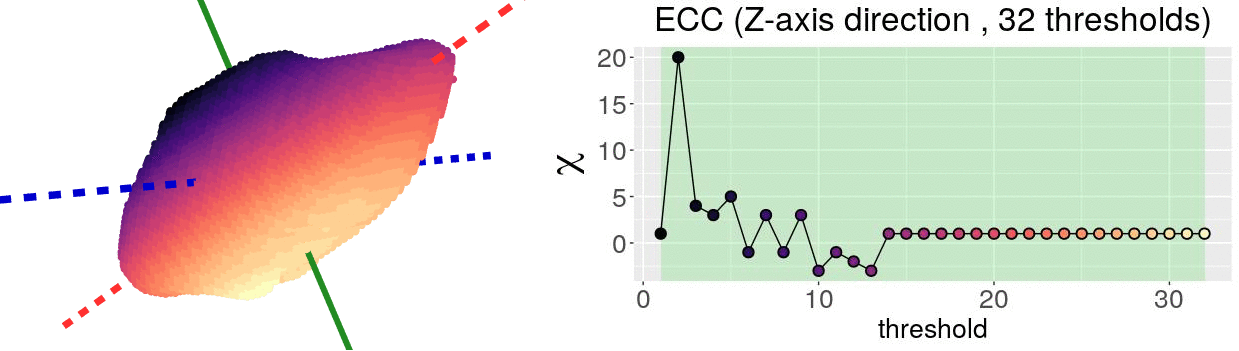
\[\chi = \#(\text{Vertices}) - \#(\text{Edges}) + \#(\text{Faces})\]
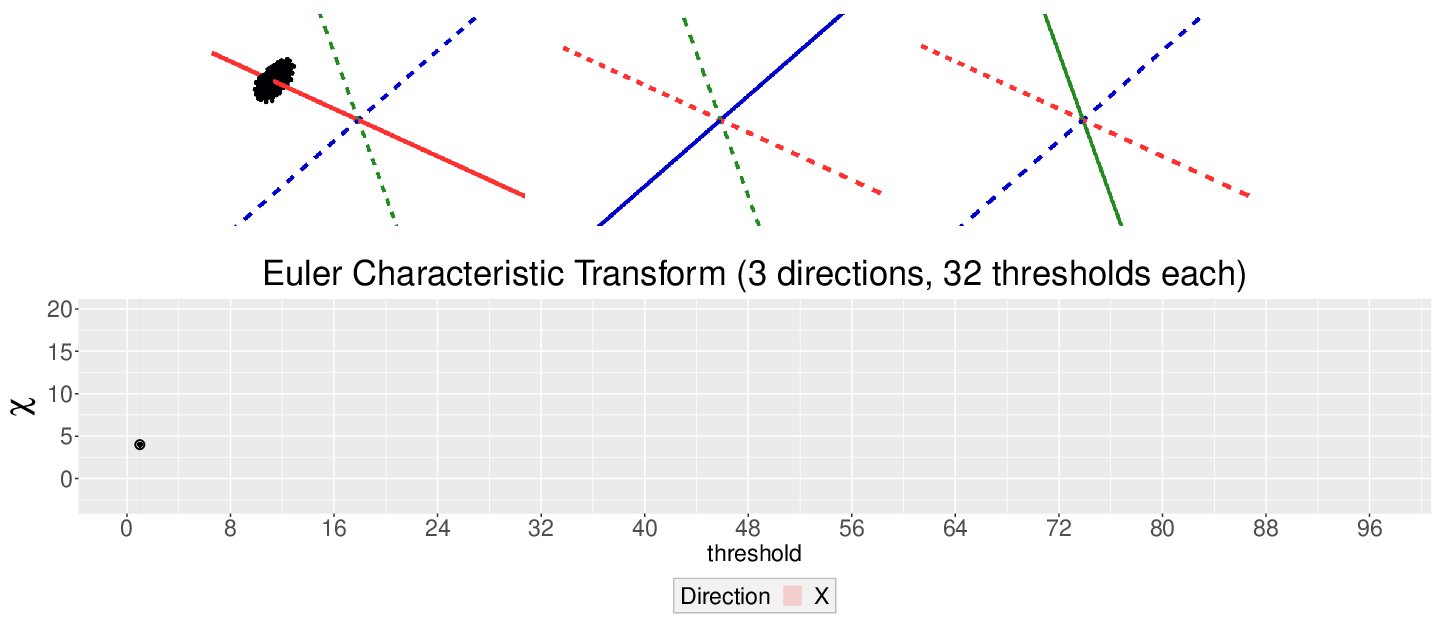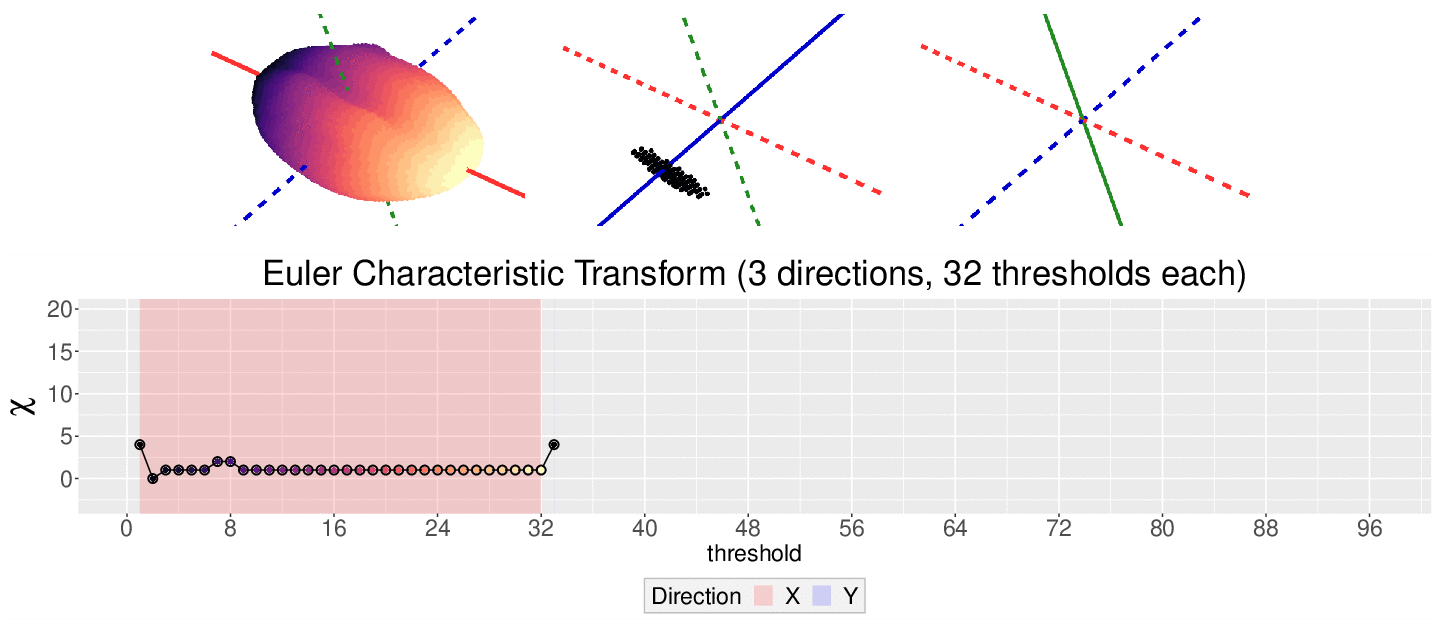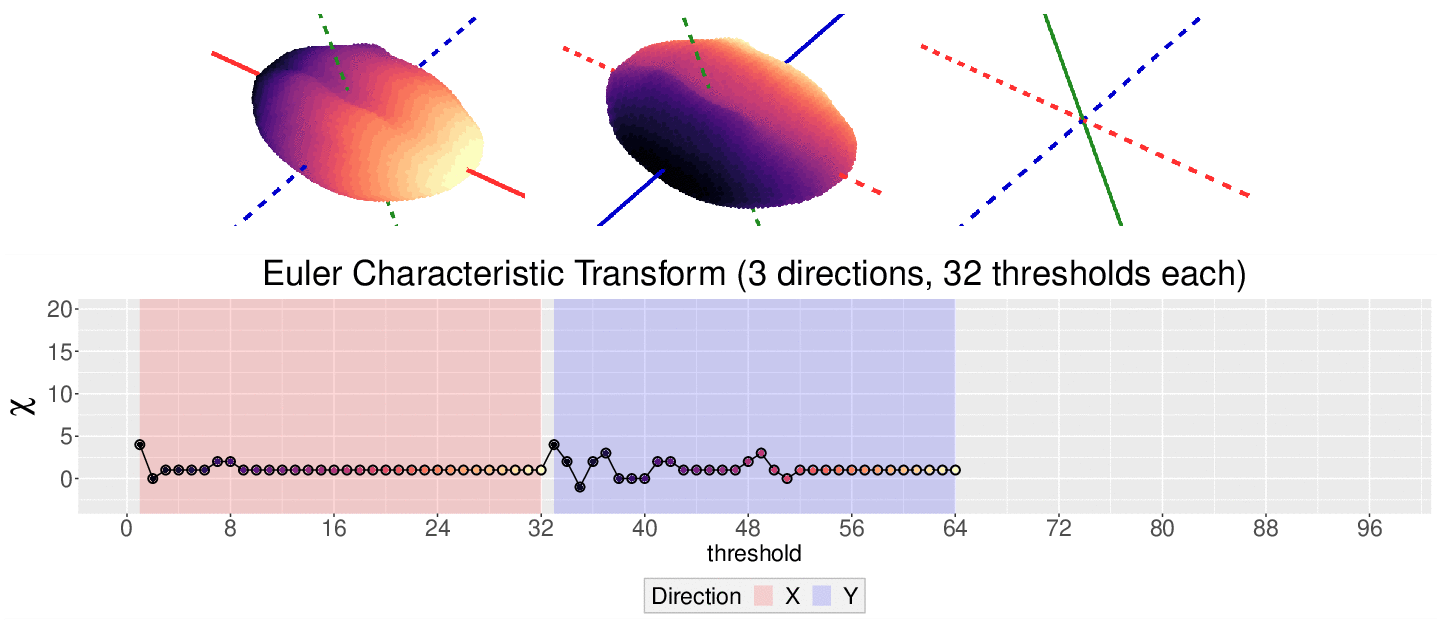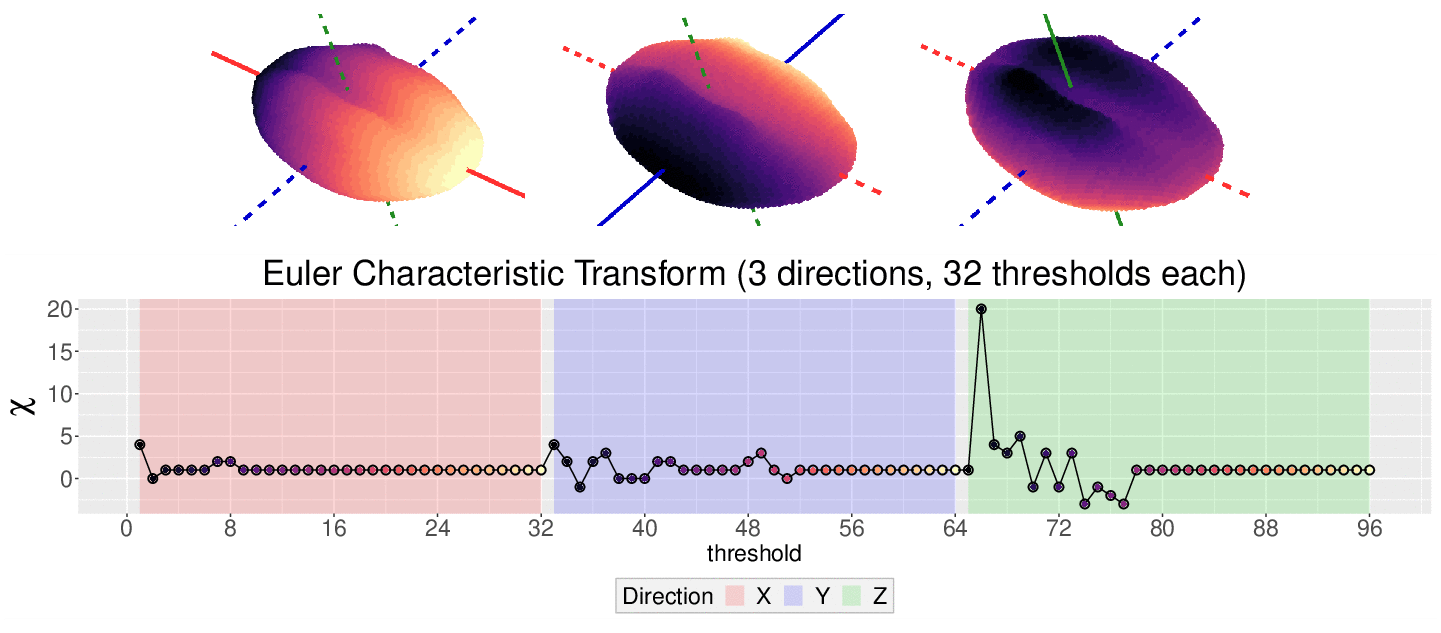
This work is supported in part by Michigan State University and the National Science Foundation Research Traineeship Program (DGE-1828149).
[1] K. Turner, S. Mukherjee, and D. M. Boyer, "Persistent homology transform for modeling shapes and surfaces," Information and Inference, vol. 3, no. 4, pp. 310–344, Dec. 2014.