class: center, middle, inverse, title-slide .title[ # Quantifying the shape of plants ] .subtitle[ ## using Topological Data Analysis ] .author[ ### <strong>Erik Amézquita</strong>, Michelle Quigley, Tim Ophelders <br> Elizabeth Munch, Dan Chitwood <br> Dan Koenig, Jacob Landis <br> - ] .institute[ ### Computational Mathematics, Science and Engineering <br> Michigan State University <br> - ] .date[ ### 2021-07-23<br> - <br> Slides available: <code>bit.ly/an21_barley</code> ] --- class: inverse # Plant morphology <div class="row"> <div class="column" style="max-width:50%"> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/oM9kAq0PBvw?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/V39K58evWlU?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> </div> <div class="column" style="max-width:50%"> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/4GBgPIEDoa0?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/qkOjHHuoUhA?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> </div> </div> --- # Topological Data Analysis <div class="row"> <div class="column" style="max-width:25%; font-size: 15px;"> <img style="padding: 25px 0 35px 0;" src="../figs/S019_L0_1.gif"> <p style="font-size: 25px; text-align: center; color: DarkRed;"> Raw Data </p> <ul> <li> X-ray CT </li> <li> Point clouds </li> <li> Time series </li> <ul> </div> <div class="column" style="max-width:40%; padding: 0 25px 0 25px; font-size: 15px;"> <img src="../figs/ecc_X.gif"> <p style="font-size: 23px; text-align: center; color: DarkRed;"> Topological Summary </p> <ul> <li> Euler Characteristic </li> <li> Persistence diagrams </li> <li> Mapper/Reeb graphs </li> <ul> </div> <div class="column" style="max-width:35%; font-size: 15px;"> <img src="../figs/svm_mds_ect.gif"> <p style="font-size: 25px; text-align: center; color: DarkRed;"> Analysis </p> <ul> <li> Statistics </li> <li> Machine learning </li> <li> Classification/prediction </li> <ul> </div> </div> --- class: inverse <div class="row"> <div class="column" style="max-width:44%"> <a href="https://kizilvest.ru/20150827-v-kizilskom-rajone-nachalas-uborochnaya-strada/" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_kizilskoye.jpg"></a> <a href="https://ipad.fas.usda.gov/highlights/2008/11/eth_25nov2008/" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_ethiopia.gif"></a> <a href="https://www.doi.org/10.1007/978-1-4419-0465-2_2168" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_historical_expansion.jpg"></a> </div> <div class="column" style="max-width:44%"> <a href="https://www.resilience.org/stories/2020-03-09/the-last-crop-before-the-desert/" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_morocco.jpg"></a> <a href="https://www.tibettravel.org/tibetan-culture/highland-barley.html" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_seed_tibet.jpg"></a> <a href="https://www.nationalgeographic.co.uk/travel/2020/05/photo-story-from-barley-fields-to-whisky-barrels-in-rural-scotland" target="_blank"><img style="padding: 0 0 0 0;" src="../figs/barley_seed_scotland_cropped.jpg"></a> </div> <div class="column" style="max-width:8%; font-size: 15px;"> <p style="text-align: center; font-size: 30px; line-height: 1em;"> <strong> Barley across the world </strong></p> <p>Kiliskoye (Chelyabinsk, Russia)</p> <p>Marchouch (Rabat, Morocco)</p> <p>Aksum (Tigray, Ethiopia)</p> <p>Salar (Tsetang, Tibet)</p> <p>Expansion of the barley. </p> <p>Turriff (Aberdeenshire, Scotland)</p> <p style="font-size:9px;line-height: 1em;">Click on any picture for more details and credits</p> </div> </div> --- # Cross Composite II experiment .pull-right[  ] --- # Cross Composite II experiment .pull-left[ 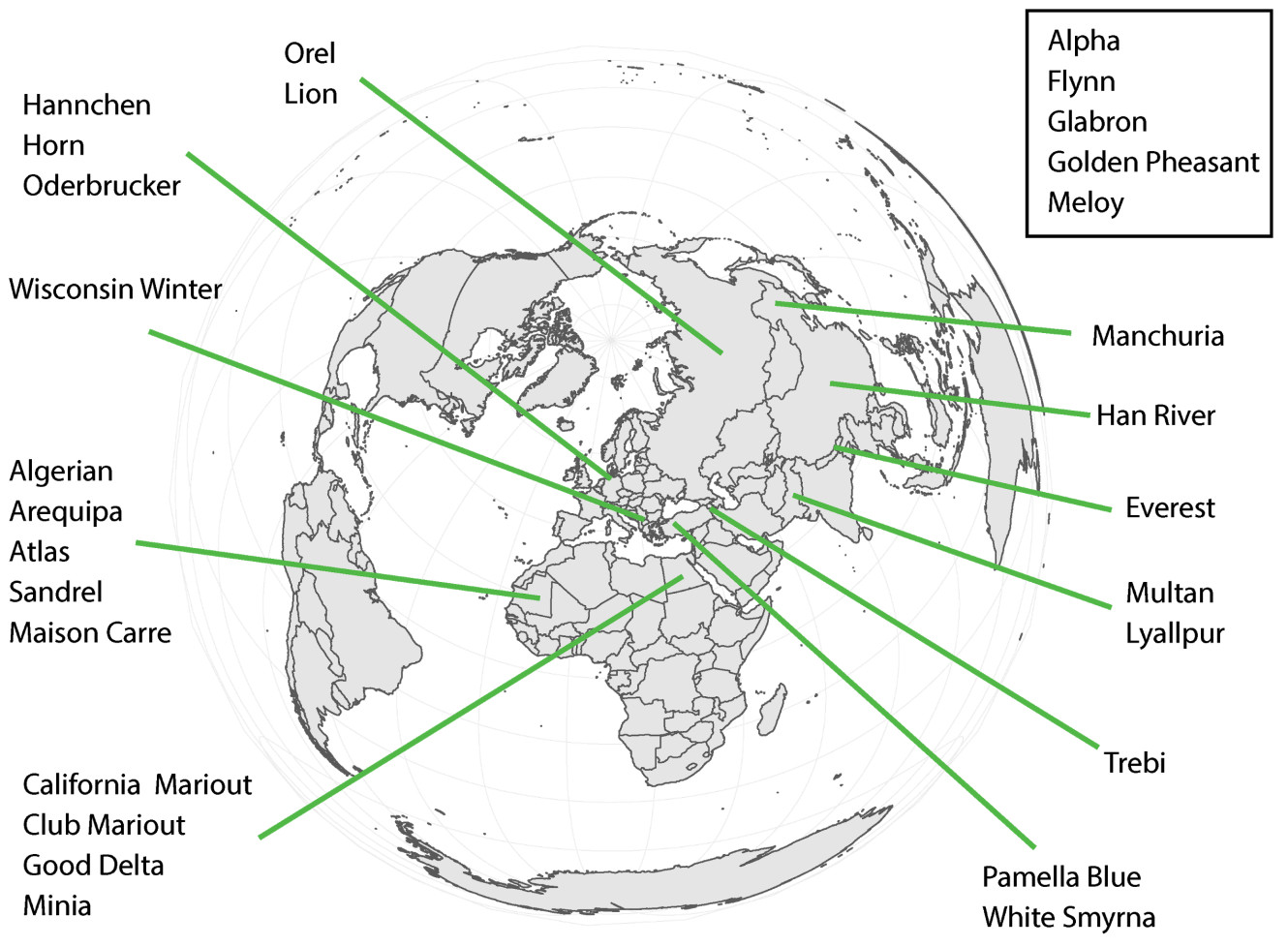 - **28 parents/accessions** `\((F_0)\)` ] .pull-right[  ] --- background-image: url("../figs/composite_cross_v_05.svg") background-size: 450px background-position: 95% 90% # Cross Composite II experiment .pull-left[  - **28 parents/accessions** `\((F_0)\)` - Do `\({28 \choose 2}=379^\dagger\)` **hybrids** `\((F_1)\)` - **Self-fertilize** the resulting 379 hybrids `\((F_2\to F_{58})\)` ] .pull-right[  ] --- # Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:51%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../figs/x3000_setup.jpg"> <p style="text-align: center;"> Proprietary X-Ray CT scan reconstruction </p> </div> </div> --- # Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:51%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../figs/x3000_setup.jpg"> <p style="text-align: center;"> Proprietary X-Ray CT scan reconstruction </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_0_original.gif"> <p style="text-align: center;"> Raw </p> </div> </div> --- # Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:51%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../figs/x3000_setup.jpg"> <p style="text-align: center;"> Proprietary X-Ray CT scan reconstruction </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_0_original.gif"> <p style="text-align: center;"> Raw </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_1_normal.gif"> <p style="text-align: center;"> Normalized </p> </div> </div> --- # Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:51%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../figs/x3000_setup.jpg"> <p style="text-align: center;"> Proprietary X-Ray CT scan reconstruction </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_0_original.gif"> <p style="text-align: center;"> Raw </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_1_normal.gif"> <p style="text-align: center;"> Normalized </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_3_denoise.gif"> <p style="text-align: center;"> Pruned </p> </div> </div> --- # Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:51%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../figs/x3000_setup.jpg"> <p style="text-align: center;"> Proprietary X-Ray CT scan reconstruction </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_0_original.gif"> <p style="text-align: center;"> Raw </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_1_normal.gif"> <p style="text-align: center;"> Normalized </p> </div> <div class="column" style="max-width:10.5%; color: Navy; font-size: 15px;"> <img src="../figs/S017_3_denoise.gif"> <p style="text-align: center;"> Pruned </p> </div> <div class="column" style="max-width:17.5%; color: Navy; font-size: 15px;"> <img src="../figs/S019_L0_1.gif"> <p style="text-align: center;"> Analysis! </p> </div> </div> -- .pull-left[ - 224 raw scans - 875 individual spikes - 3 generations: `\(F_0, F_{18}, F_{58}\)` - 38,000 clean seeds ] -- .pull-right[  ] --- ## Image processing → Traditional descriptors .pull-left[ - Length - Width - Height - Surface area - Volume  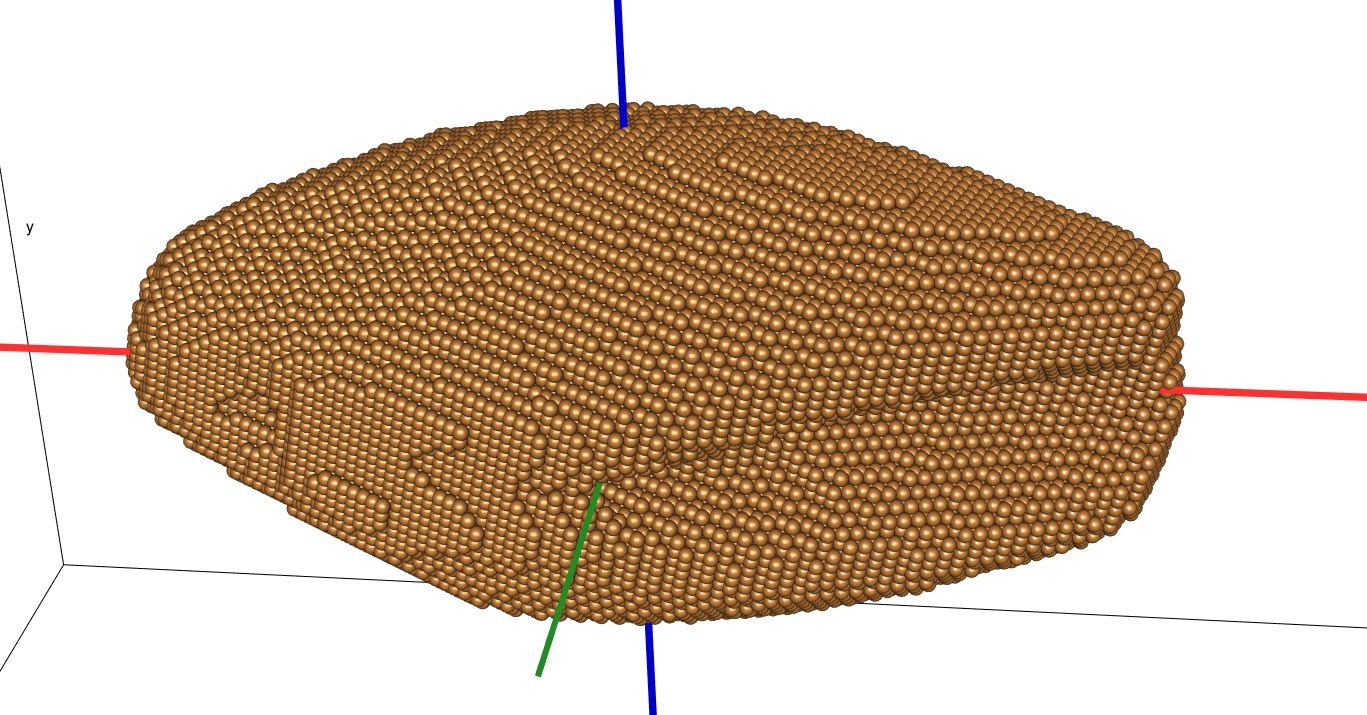 ] -- .pull-right[ 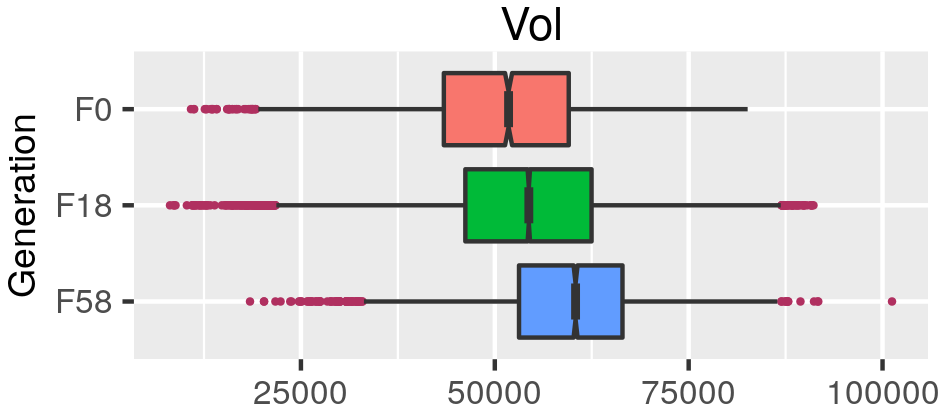 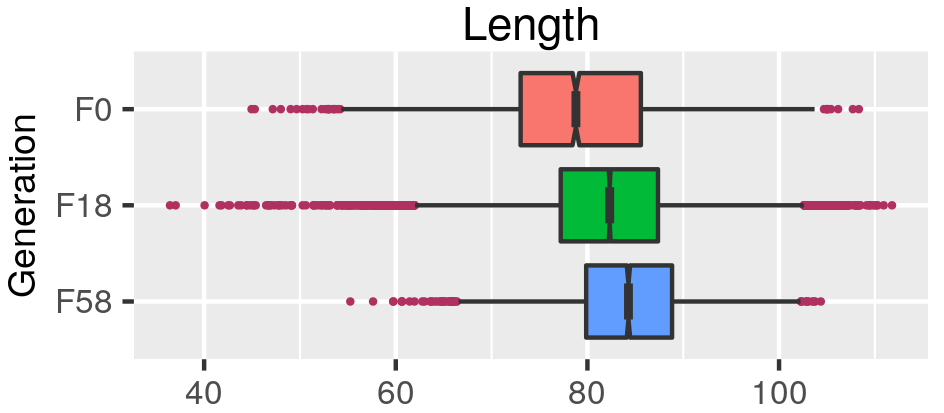  ] --- # Topology: The Euler characteristic `\(\chi\)` `$$\chi = \#(\text{Vertices}) - \#(\text{Edges}) + \#(\text{Faces}).$$` <img src="../../tda/figs/euler_characteristic_2.png" width="400" style="display: block; margin: auto;" /> -- - Summarize **topological features** with the Euler-Poincaré formula `$$\chi = \#(\text{Connected Components}) - \#(\text{Loops}) + \#(\text{Voids}).$$` -- - The Euler characteristic is a **topological invariant**. --- # Euler Characteristic Curve (ECC) - Consider a cubical complex `\(X\subset\mathbb{R}^d\)` - And a unit-length direction `\(\nu\in S^{d-1}\)` -- - And the subcomplex containing all cubical cells below height `\(h\)` in the direction `\(\nu\)` `$$X(\nu)_h =\{\Delta \in X\::\:\langle x,\nu\rangle\leq h\text{ for all }x\in\Delta\}$$` -- - The Euler Characteristic Curve (ECC) of direction `\(\nu\)` is defined as the sequence `$$\{\chi(X(\nu)_h)\}_{h\in\mathbb{R}}$$` --- background-image: url("../figs/ecc_ver2.gif") background-size: 750px background-position: 50% 90% # Euler Characteristic Curve (ECC) - Consider a cubical complex `\(X\subset\mathbb{R}^d\)` - And a unit-length direction `\(\nu\in S^{d-1}\)` - And the subcomplex containing all cubical cells below height `\(h\)` in the direction `\(\nu\)` `$$X(\nu)_h =\{\Delta \in X\::\:\langle x,\nu\rangle\leq h\text{ for all }x\in\Delta\}$$` - The Euler Characteristic Curve (ECC) of direction `\(\nu\)` is defined as the sequence `$$\{\chi(X(\nu)_h)\}_{h\in\mathbb{R}}$$` --- # Euler Characteristic Transform (ECT) - Repeat and concatenate for all possible directions. - More formally, the ECT can be thought as a function $$ `\begin{split} ECT(X):\; & S^{d-1} \to \mathbb{Z}^{\mathbb{R}}\\ &\nu\mapsto\{\chi(X(\nu)_h)\}_{h\in\mathbb{R}}. \end{split}` $$ --- background-image: url("../figs/ect_ver2.gif") background-size: 800px background-position: 50% 88% # Euler Characteristic Transform (ECT) - Repeat and concatenate for all possible directions. - More formally, the ECT can be thought as a function $$ `\begin{split} ECT(X):\; & S^{d-1} \to \mathbb{Z}^{\mathbb{R}}\\ &\nu\mapsto\{\chi(X(\nu)_h)\}_{h\in\mathbb{R}}. \end{split}` $$ --- # Why choose the ECT? -- - Easy to compute: a quick alternating sum. -- [**Theorem _(Turner, Mukherjee, Boyer 2014)_**](https://doi.org/10.1093/imaiai/iau011): The ECT is injective for finite simplicial complexes in 3D. [**Theorem _(ibid)_**](https://arxiv.org/abs/1310.1030): The ECT is a sufficient statistic for finite simplicial complexes in 3D. -- *Translation:* - Given all the (infinite) ECCs corresponding to all possible directions, - *Different* simplicial complexes correspond to *different* ECTs. - The ECT effectively summarizes all possible information related to shape. -- There is math research on computationally efficient reconstruction algorithms for 3D shapes ([Turner, Curry](https://arxiv.org/abs/1805.09782), [Fasy](https://arxiv.org/abs/1912.12759), [Ghrist](https://doi.org/10.1007/s41468-018-0017-1)) but this remains elusive. --- background-image: url("../figs/S012_L2_Blue_33.png") background-size: 150px background-position: 99% 50% # Game plan - **Goal:** Classify 28 barley parental genotypes using solely grain morphology information. - **3121** grains in total -- .pull-left[ <img src="../figs/pole_directions_p7_m12_crop.jpg" width="150" style="display: block; margin: auto;" /> ] .pull-right[ - 158 directions - 8 thresholds per direction - Every seed is associated a `\(158\times8=1264\)`-dimensional vector - Dimensions reduced with UMAP ] -- - Compare **3** sets of morphological descriptors Descriptor | No. of descriptors -----------|-------------------- Traditional | 11 Topological (ECT → UMAP) | ~~1264~~ → 12 Combined (Trad ⊕ Topo) | 23 - Sample randomly 75/25 training/testing sample for each genotype. - Repeat the sampling and SVM computation 100 and consider the average. --- # Classification of 28 lines with SVM <style type="text/css"> .tg {border-collapse:collapse;border-color:#93a1a1;border-spacing:0;margin:0px auto;} .tg td{background-color:#fdf6e3;border-bottom-width:1px;border-color:#93a1a1;border-style:solid;border-top-width:1px; border-width:0px;color:#002b36;font-family:Arial, sans-serif;font-size:14px;overflow:hidden;padding:10px 5px; word-break:normal;} .tg th{background-color:#657b83;border-bottom-width:1px;border-color:#93a1a1;border-style:solid;border-top-width:1px; border-width:0px;color:#fdf6e3;font-family:Arial, sans-serif;font-size:14px;font-weight:normal;overflow:hidden; padding:10px 5px;word-break:normal;} .tg .tg-2bhk{background-color:#eee8d5;border-color:inherit;text-align:left;vertical-align:top} .tg .tg-0pky{border-color:inherit;text-align:left;vertical-align:top} .tg .tg-gyvr{background-color:#eee8d5;border-color:inherit;font-size:100%;text-align:left;vertical-align:top} </style> <table class="tg"> <thead> <tr> <th class="tg-0pky">Shape descriptors</th> <th class="tg-0pky">No. of descriptors</th> <th class="tg-0pky">Precision</th> <th class="tg-0pky">Recall</th> <th class="tg-0pky">F1</th> </tr> </thead> <tbody> <tr> <td class="tg-2bhk">Traditional</td> <td class="tg-2bhk">11</td> <td class="tg-2bhk">0.57 ± 0.058</td> <td class="tg-2bhk">0.56 ± 0.019</td> <td class="tg-2bhk">0.55 ± 0.019</td> </tr> <tr> <td class="tg-0pky">Topological</td> <td class="tg-0pky">12</td> <td class="tg-0pky">0.75 ± 0.047</td> <td class="tg-0pky">0.75 ± 0.016</td> <td class="tg-0pky">0.74 ± 0.016</td> </tr> <tr> <td class="tg-2bhk">Combined</td> <td class="tg-2bhk">23</td> <td class="tg-2bhk">0.87 ± 0.031</td> <td class="tg-2bhk">0.86 ± 0.010</td> <td class="tg-2bhk">0.86 ± 0.010</td> </tr> </tbody> </table> <img src="../figs/avg_f1_combined_158_16_12_umap_horz.png" width="700" style="display: block; margin: auto;" /> --- # Hidden topological shape information .pull-left[ - Analysis of variance to determine the most discerning directions and slices/thresholds. - The top crease on the seed is highly descriptive! <img src="../figs/kruskal_wallis_topo_summary.jpg" width="300" style="display: block; margin: auto;" /> ] -- .pull-right[ <img src="../figs/discerning_directions.png" width="225" style="display: block; margin: auto;" />  ] --- # Into semi-supervised territory - Train an SVM with 100% of the founders `\((F_0)\)` - Classify the progeny `\((F_{18}\text{ and }F_{58})\)` to detect genotype enrichment .pull-left[  ] .pull-right[  ] --- class: right, bottom, inverse background-image: url("../figs/acknowledgments.jpg") background-size: 1000px background-position: 50% 40% Grab these slides at [`bit.ly/an21_barley`](http://bit.ly/an21_barley)