class: center, middle, inverse, title-slide .title[ # Si la vida te da limones ] .subtitle[ ## modela la distribución de sus glándulas y aroma ] .author[ ### <strong>Erik Amézquita</strong>, Michelle Quigley, Tim Ophelders <br> Danelle Seymour, Elizabeth Munch, Dan Chitwood <br> - ] .institute[ ### Division of Plant Sciences & Technology<br>University of Missouri<br> - ] .date[ ### 2023-12-13<br> - <br> Publicado en <a href="https://doi.org/10.1002/ppp3.10333">Plants, People, Planet (2022)</a> ] --- background-image: url("../../img/endlessforms.png") background-size: 150px background-position: 89% 7% class: inverse # When life gives you lemons... <div class="row"> <div class="column" style="max-width:50%"> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/khRS5AuleIM?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/HAIaektvV3Q?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> </div> <div class="column" style="max-width:50%"> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/Xo2HULkkm3s?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> <iframe width="375" height="210" src="https://www.youtube-nocookie.com/embed/EwTwSNrBuIo?controls=0" frameborder="0" allow="accelerometer; autoplay; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe> </div> </div> <p style="font-size: 24px; text-align: right; font-family: 'Yanone Kaffeesatz'">More X-ray CT 3D scans at <a href="https://www.youtube.com/@endlessforms6756">youtube.com/@endlessforms6756</a></p> --- background-image: url("../../demat/figs/fam9_3.png") background-size: 100px background-position: 98% 2% # From MX to MI to MO at MU .left-column[  ] .right-column[ - 2013 - 2018 : Math @ at the Universidad de Guanajuato and CIMAT (Mexico). - 2016 - 2018 : Bachelor thesis: **Math + Archaeology**. Use TDA to quantify and classify the shape of pre-Columbian masks found in the Templo Mayor in Mexico City. - 2018 - 2023 : CMSE @ Michigan State University. **Came for the math. Stayed for the plants.** - 2023 - present : PFFIE Postdoc Fellow @ Division of Plant Sciences & Department of Mathematics at Mizzou ]  --- background-image: url("../../img/primary-logo-forestgreen.svg") background-size: 100px background-position: 98% 2% # My main past projects <div class="row" style="font-family: 'Yanone Kaffeesatz'; font-size:25px;"> <div class="column" style="max-width:25%"> <p style="text-align: center;">The shape of<br>adaptability</p> <img style="padding: 0 40px;" src="../../barley/figs/S017_L3_1.gif"></img> <img style="padding: 0 30px;" src="../../barley/figs/ecc_X.gif"></img> <p style="text-align: center; font-size:20px;">Topological Data Analysis<br>Euler Characteristic Transform</p> </div> <div class="column" style="max-width:25%"> <p style="text-align: center;">The shape of<br>development</p> <img style="padding: 0 25px;" src="../../citrus/figs/SR01_L01_black_exocarp.gif"></img> <img style="padding: 0 10px;" src="../../citrus/figs/SW03_CRC1241-B_12B-4-3_L00_lambproj_v.jpg"></img> <p style="text-align: center; font-size:20px">Ellipsoidal modeling<br>Directional statistics</p> </div> <div class="column" style="max-width:25%"> <p style="text-align: center;">The shape of<br>breeding</p> <img style="padding: 0;" src="../../walnuts/figs/2014SBa_R5_T81_meat_2.gif"></img> <img style="padding: 0;" src="../../walnuts/figs/kernel_volume_vs_nut_volume.png"></img> <p style="text-align: center; font-size:20px">Allometry of multiple tissues<br>Convexity indices</p> </div> <div class="column" style="max-width:25%"> <p style="text-align: center;">The shape of<br>omics</p> <img style="padding: 0;" src="../../nasrin/figs/fpkm_raw_3.png"></img> <img style="padding: 0 10px;" src="../../nasrin/figs/lung_t0_meancorr_eps7.0e+02_r40_g40.png"></img> <img style="padding: 0 10px;" src="../../nasrin/figs/lung_tpm_maxcorr_eps7.0e+04_r40_g30.png"></img> <p style="text-align: center; font-size:20px">Mapper<br>Spectral analysis</p> </div> </div> --- # Roadmap for today .left-column[   ] .right-column[ 1. Brief overview of citrus biology and cultural history 1. Experimental setup and quick results 1. Ellipsoidal approximation and directional statistics The last 20 slides are a crash course into Topological Data Analysis. Will **not** get there unless there is ample time. ] --- class: inverse, middle, center # More than just oranges <img src="../crc_pics/south_coast_field_station_citron.jpg" width="400" style="display: block; margin: auto;" /> --- ## Think of citrus as lego blocks 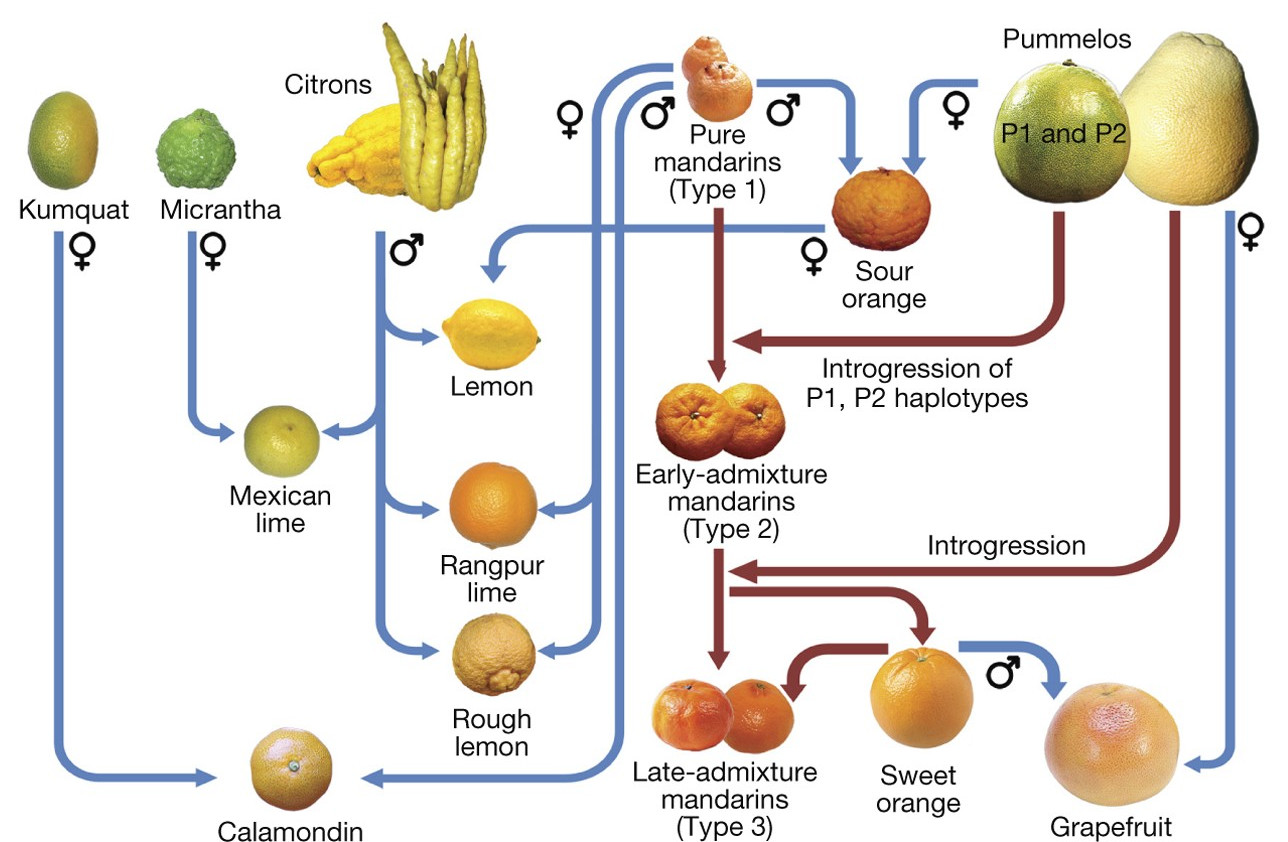 <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://doi.org/10.1038/nature25447">Wu <em>et al.</em> (2018)</a></p> --- background-image: url("../crc_pics/cvc3.jpg") background-size: 350px background-position: 100% 0% ## Citrus originated 8M* years ago Originated in South and South East Asia, and Australia. <img src="../figs/precursor_citrus_geo_distribution.jpg" width="570" style="display: block; margin: auto;" /> <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://doi.org/10.1038/nature25447">Wu <em>et al.</em> (2018)</a></p> --- background-image: url("../crc_pics/cvc2.jpg") background-size: 850px background-position: 50% 40% ## Genetic distances between citrus <img src="../figs/../figs/precursor_citrus_phylogenetic.jpg" width="550" style="display: block; margin: auto auto auto 0;" /> <p style="font-size: 8px; text-align: center; color: Grey;"> Credits: <a href="https://doi.org/10.1038/nature25447">Wu <em>et al.</em> (2018)</a></p> 10* _fundamental_ citrus gave rise to all citrus diversity we see today --- ## Oil glands are closely linked to fruit development .pull-left[  <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://www.boredpanda.com/life-cycles-pics/">BoredPanda</a></p> Developing cycle of a lemon ] .pull-right[ <iframe width="560" height="300" src="https://static-movie-usa.glencoesoftware.com/webm/10.1073/956/d916befc88029defb1ecef6c4a2fd83db89428d9/pnas.1720809115.sm02.webm" frameborder="0" allowfullscreen></iframe> <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://doi.org/10.1073/pnas.1720809115">Smith <em>et al.</em> (2018)</a></p> - Cross-sectional view of a navel orange peel bending to the point of jetting. - Huge perfume and food industry behind essential oils ] --- class: inverse, middle, center # Experimental setup ## And some quick results <img src="../crc_pics/citrus_webberii.jpg" width="400" style="display: block; margin: auto;" /> --- ## Raw Data: X-rays → Image Processing <div class="row"> <div class="column" style="max-width:38%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../../citrus/crc_pics/crc_diversity.jpg"> <p style="text-align: center;"> UCR Collaboration </p> </div> <div class="column" style="max-width:38%; color: Navy; font-size: 15px;"> <img style="padding: 2px 0 2px 0;" src="../../citrus/crc_pics/citrus_xrayct_scanning.jpeg"> <p style="text-align: center;"> 3D X-Ray CT scan </p> </div> <div class="column" style="max-width:23%; color: Navy; font-size: 15px;"> <img src="../../citrus/figs/SR01_CRC3289_12B-19-9_L01_raw.gif"> <p style="text-align: center;"> Raw </p> </div> </div> <div class="row" style="margin: 0 auto;"> <div class="column" style="max-width:20%; color: Navy; font-size: 15px;"> <img src="../figs/SR01_L01_black_spine.gif"> <p style="text-align: center;"> Spine </p> </div> <div class="column" style="max-width:20%; color: Navy; font-size: 15px;"> <img src="../figs/SR01_L01_black_endocarp.gif"> <p style="text-align: center;"> Endocarp </p> </div> <div class="column" style="max-width:20%; color: Navy; font-size: 15px;"> <img src="../figs/SR01_L01_black_rind.gif"> <p style="text-align: center;"> Rind </p> </div> <div class="column" style="max-width:20%; color: Navy; font-size: 15px;"> <img src="../figs/SR01_L01_black_exocarp.gif"> <p style="text-align: center;"> Exocarp </p> </div> <div class="column" style="max-width:20%; color: Navy; font-size: 15px;"> <img src="../figs/SR01_L01_black_oil_glands.gif"> <p style="text-align: center;"> Oil glands</p> </div> </div> --- ## The centers will be just fine .pull-left[ 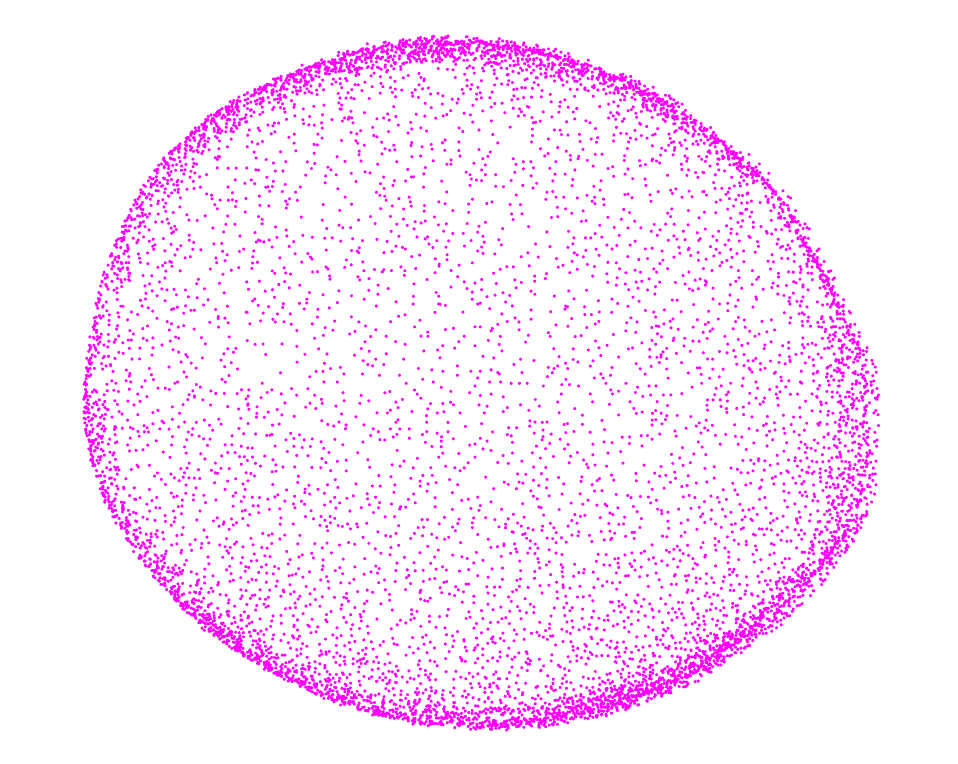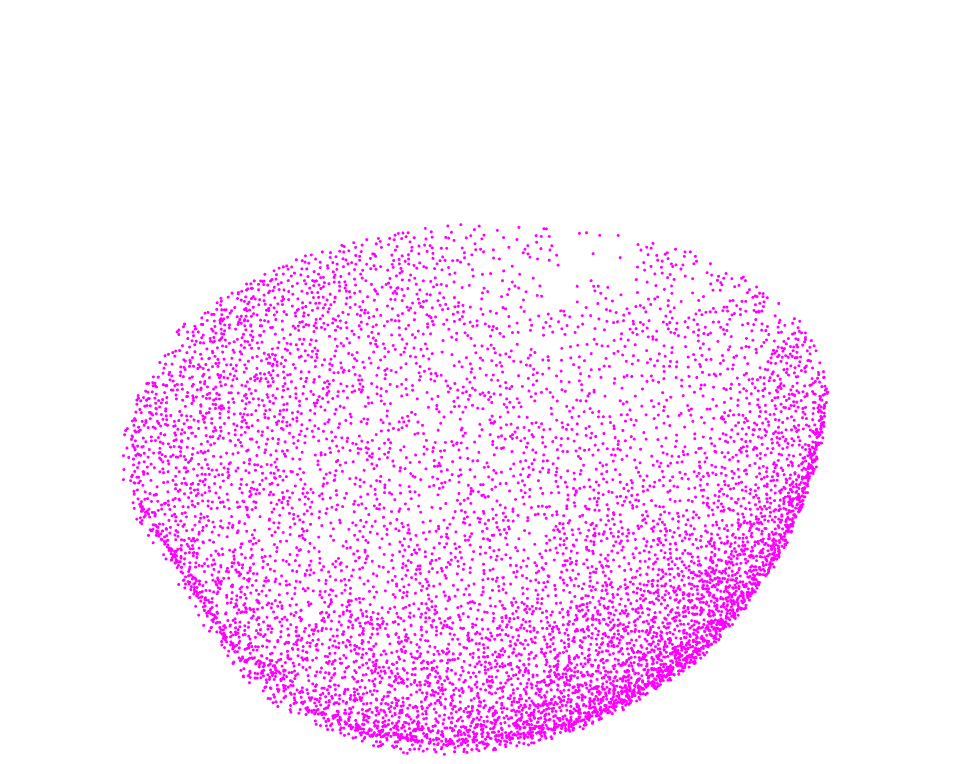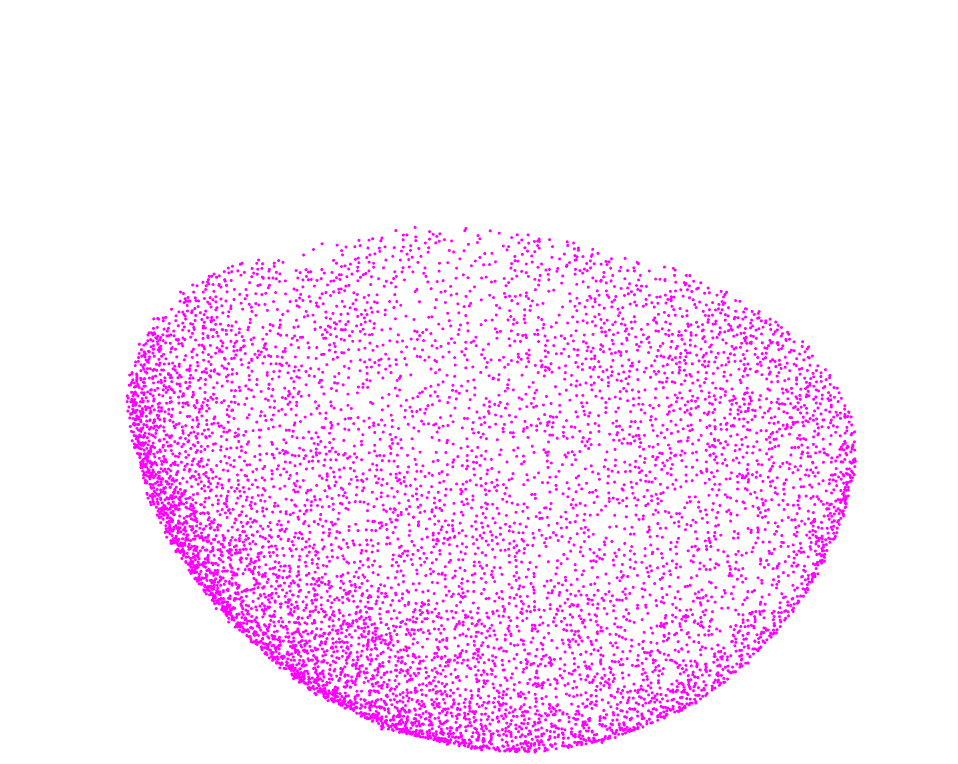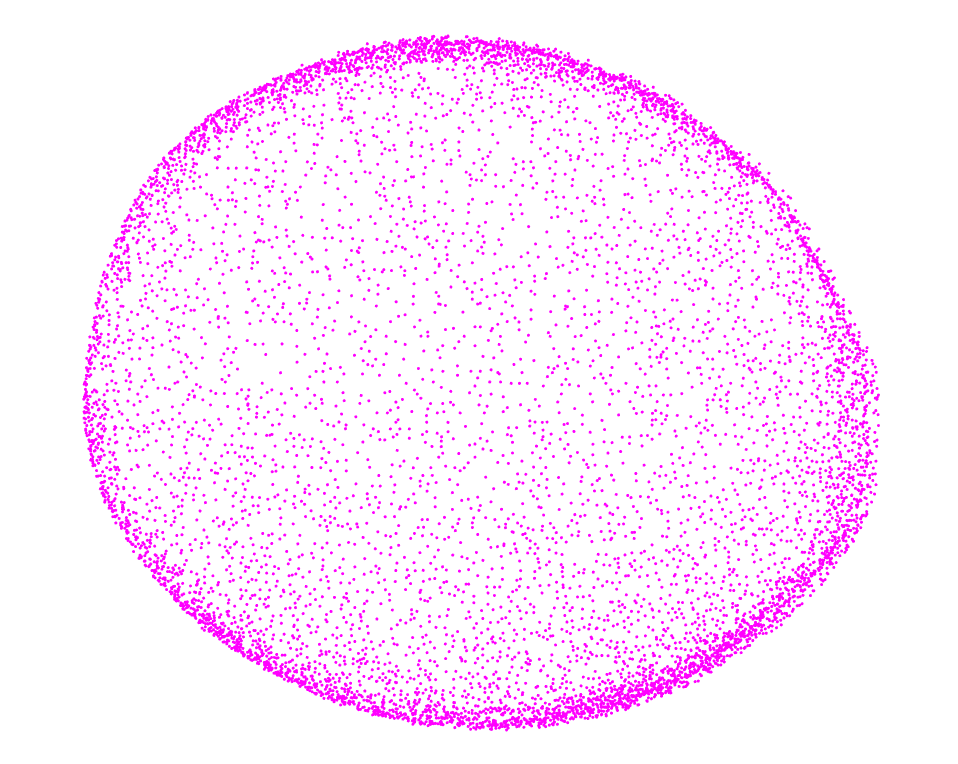 - 58 species and 163 raw scans - About 6K→20K individual oil glands detected - Number in the right ballpark - We have a point cloud in `\(\mathbb{R}^3\)`. ] .pull-right[  <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://doi.org/10.1006/anbo.2001.1546">Knight <em>et al.</em> (2001)</a></p> ] --- # Allometry + Linear relationship between size-related measurements. <div class="row" style="margin: 0 auto;"> <div class="column" style="max-width:49%; color: Navy; font-size: 15px;"> <img src="../figs/exocarp_vs_endocarp.jpg"> <img src="../figs/whole_vs_oil_glands.jpg"> </div> <div class="column" style="max-width:49%; color: Navy; font-size: 15px;"> <img src="../figs/whole_vs_rind.jpg"> <img src="../figs/oil_glands_vs_num_glands.jpg"> </div> </div> + Total number of glands seems to be decoupled --- ## Average distance from each gland to its `\(k\)`-th nearest neighbor  + `\(\text{Avg.dist}(k) = \sqrt{Mk + B}\)` + Oil glands might be distributing themselves following **normal diffusion** mechanics + The **outliers** for citrus groups usually correspond to hybrids. --- class: inverse, center, middle # Citrus are quite spherical ## Ellipsoidal to be more precise ## Directional statistics to compare fruit surfaces <img src="../crc_pics/nagami_kumquat.jpg" width="400" style="display: block; margin: auto;" /> --- background-image: url("../ellipsoids/Li_and_Griffiths_2004.png") background-size: 250px background-position: 95% 3% ## Best ellipsoid fitting — least squares - A general quadratic surface is defined by the equation `$$\eqalignno{ & ax^{2}+by^{2}+cz^{2}+2fxy+2gyz+2hzy\ \ \ \ \ \ \ \ \ &\hbox{(1)}\cr &+2px+2qy+2rz+d=0.}$$` With our observations `\(\{(x_i,y_i,z_i)\}_i\)`, $$ `\begin{pmatrix} x_1^2 & y_1^2 & z_1^2 & 2x_1y_1 & 2y_1z_1 & 2x_1z_1 & x_1 & y_1 & z_1 & 1\\ x_2^2 & y_2^2 & z_2^2 & 2x_2y_2 & 2y_2z_2 & 2x_2z_2 & x_2 & y_2 & z_2 & 1\\ \vdots& \vdots& \vdots& \vdots & \vdots & \vdots & \vdots & \vdots & \vdots \\ x_n^2 & y_n^2 & z_n^2 & 2x_ny_n & 2y_nz_n & 2x_nz_n & x_n & y_n & z_n & 1 \end{pmatrix}` `\begin{pmatrix} a \\ b \\ \vdots \\ d \end{pmatrix}` = `\begin{pmatrix} 0 \\ 0 \\ \vdots \\ 0 \end{pmatrix}` $$ or $$ \mathbf{D}\mathbf{v} = 0 $$ The solution to the system above can be obtained via Lagrange multipliers `$$\min_{\mathbf{v}\in\mathbb{R}^{10}}\left\|\mathbf{D}\mathbf{v}\right\|^2, \quad \mathrm{s.t.}\; \text{ some ellipsoid-specific conditions hold}$$` --- ## Model the whole fruit as an ellipsoid .pull-left[   ] .pull-right[   ] --- background-image: url("../figs/lambert_equal_area_N.gif") background-size: 150px background-position: 98% 1% ## Citrus modeling: Sour orange  <img src="../figs/SR01_CRC3289_12B-19-9_L00_geocentric_lazi.jpg" width="550" style="display: block; margin: auto;" /> --- background-image: url("../figs/lambert_equal_area_N.gif") background-size: 150px background-position: 98% 1% ## Citrus modeling: Pummelo  <img src="../figs/P01A_CRC3781_12A-31-9_L00_geocentric_lazi.jpg" width="550" style="display: block; margin: auto;" /> --- background-image: url("../figs/lambert_equal_area_N.gif") background-size: 150px background-position: 98% 1% ## Citrus modeling: Mandarin  <img src="../figs/M01_CRC3226_12B-27-1_L00_geocentric_lazi.jpg" width="550" style="display: block; margin: auto;" /> --- background-image: url("../figs/lambert_equal_area_N.gif") background-size: 150px background-position: 98% 1% ## Citrus modeling: Kumquat  <img src="../figs/WR02_CRC3877_12B-44-13_L00_geocentric_lazi.jpg" width="550" style="display: block; margin: auto;" /> --- background-image: url("../../citrus/figs/lambert_equal_area_N.gif") background-size: 150px background-position: 98% 1% ## Citrus modeling: Finger lime  <img src="../figs/WR08_CRC3661_18B-16-5_L00_geocentric_lazi.jpg" width="550" style="display: block; margin: auto;" /> --- background-image: url("../ellipsoids/GarciaPortugues_etal_2020a.png") background-size: 200px background-position: 95% 3% # Can we say something more?  --- # Uniform and rotationally symmetric spherical distributions  --- class: inverse, center, middle # But then, what distribution do oil glands follow? ## Spherical Kernel Density Estimates FTW <img src="../crc_pics/finger_lime.jpg" width="400" style="display: block; margin: auto;" /> --- # Kernel Density Estimators  <p style="font-size: 8px; text-align: right; color: Grey;"> Credits: <a href="https://www.statsmodels.org/dev/examples/notebooks/generated/kernel_density.html">Statsmodels</a></p> - The choice of bandwidth is crucial - Large bandwidths tend to oversmooth - Small bandwidths tend to overfit - Plenty of literature on `\(\mathbb{R}\)` on how to choose an adequate value --- background-image: url("../ellipsoids/GarciaPortugues_2013.png") background-size: 300px background-position: 95% 3% # Spherical KDE  - Oil glands of a papeda - Red arrows indicate the most significant gradient values for the density function - Concentration parameter `\(k\)` is chosen to be optimal --- background-image: url("../ellipsoids/Vuollo_and_Holmstrom_2018.png") background-size: 300px background-position: 95% 3% # More exploratory analysis ## SphereSiZer: Pummelo  --- # Side view of a sweet orange  --- # Side view of a mandarin  --- # Side view of a kumquat  --- # Side view of a finger lime  --- background-image: url("../../citrus/figs/ppp3.png") background-size: 225px background-position: 95% 3% # Summary .left-column[ <img src="../../citrus/crc_pics/citrus_amblycarpa.jpg" width="150" style="display: block; margin: auto;" /><img src="../../citrus/figs/SR01_L01_black_exocarp.gif" width="150" style="display: block; margin: auto;" /><img src="../../citrus/figs/SW03_CRC1241-B_12B-4-3_L00_lambproj_v.jpg" width="150" style="display: block; margin: auto;" /> ] .right-column[ - It is possible to **compare** ~~apples~~ limes to oranges, despite diversity in shapes and sizes. - We can define the non-parametrical **distribution** of oil glands for most of the citrus. - **First** time citrus shape has been examined using X-ray CT scans. - AFAIK, this is also the **first** time directional statistics have been **applied** to plant biology. ] --- background-image: url("../../img/phd_institutional_logos.jpg") background-size: 500px background-position: 95% 5% class: inverse # Thank you! <div class="row"> <div class="column" style="max-width:23%; font-size: 15px;"> <img style="padding: 0 0 0 0;" src="https://i1.rgstatic.net/ii/profile.image/607374528233472-1521820780707_Q128/Elizabeth-Munch.jpg"> <p style="text-align: center;">Liz Munch (MSU)</p> <img style="padding: 0 0 0 0;" src="https://alga.win.tue.nl/images/staff/tim.jpg"> <p style="text-align: center;">Tim Ophelders (Utrecht)</p> </div> <div class="column" style="max-width:23%; font-size: 15px;"> <img style="padding: 0 0 0 0;" src="https://www.canr.msu.edu/contentAsset/image/9ae9777d-157c-46e6-9f12-d062ad35671e/fileAsset/filter/Jpeg,Resize,Crop/jpeg_q/80/resize_w/400/crop_x/0/crop_y/45/crop_w/400/crop_h/400"> <p style="text-align: center;">Dan Chitwood<br>(MSU)</p> <img style="padding: 0 0 0 0;" src="https://i1.rgstatic.net/ii/profile.image/926632407748609-1597937790454_Q128/Michelle-Quigley.jpg"> <p style="text-align: center;">Michelle Quigley<br>(MSU)</p> </div> <div class="column" style="max-width:16%; font-size: 15px;"> <img style="padding: 0 0 0 0;" src="https://iigb.ucr.edu/sites/g/files/rcwecm5716/files/styles/scale_225/public/Seymour.jpg?itok=pnUK_orh"> <p style="text-align: center;">Danelle Seymour<br>(UC Riverside)</p> <img style="padding: 0 0 0 0;" src="../figs/SR01_L01_black_exocarp.gif"> </div> <div class="column" style="width:10%; font-size: 24px;"> </div> <div class="column" style="max-width:30%; font-size: 24px; line-height:1.25"> <p style="text-align: center;"><strong>Email</strong></p> <p style="text-align: center; color: Blue">eah4d@missouri.edu</p> <p style="text-align: center;"><strong>Website and slides</strong></p> <p style="text-align: center; color: Blue">ejamezquita.github.io</p> <img style="padding: 0 0 0 0;" src="../figs/ppp3.png"> </div> </div> --- class: inverse, center, middle # Do I still have time? <img src="../crc_pics/cvc12.jpg" width="300" style="display: block; margin: auto;" /> --- class: inverse, center, middle # Future topological directions ## A crash course in Topological Data Analysis <img src="../crc_pics/interdonato_lemon.jpg" width="300" style="display: block; margin: auto;" /> --- # 1st TDA Ingredient: Complexes - Think the data as a collection of elementary building blocks ( _simplices_ ) Vertices | Edges | Faces | Tetrahedra ---------|-------|-------|------- 0-dim | 1-dim | 2-dim | 3-dim - A collection of cells is a _simplicial complex_ - Count the number of topologically invariant features ( _holes_ ): Connected components | Loops | Voids ---------------------|-------|------- 0-dim | 1-dim | 2-dim - Example with 3 connected components, 1 loop, 1 voids <img src="../../tda/figs/complex-good.svg" width="350" style="display: block; margin: auto;" /> --- # 2nd TDA Ingredient: Filters - Each cell is assigned a real value which defines how the complex is constructed. - Observe how the number of topological features change as the complex grows. .pull-left[ <img src="../../barley/figs/eigcurv_filter.gif" width="250px" style="display: block; margin: auto;" /><img src="../../barley/figs/gaussian_density_filter.gif" width="250px" style="display: block; margin: auto;" /> ] .pull-right[ <img src="../../barley/figs/eccentricity_filter.gif" width="250px" style="display: block; margin: auto;" /><img src="../../barley/figs/vrips_ver2.gif" width="250px" style="display: block; margin: auto;" /> ] --- background-image: url("../../tda/figs/vr1_00.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_01.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_02.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_03.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_04.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_05.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_06.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/vr1_07.svg") background-size: 475px background-position: 50% 90% # A toy Vietoris-Rips filtration example - Consider a collection of vertices `\(\{v_\lambda\}_{\lambda\in\Lambda}\)` and `\(r\geq0\)`. - The subset `\(\{v_0,\ldots,v_n\}\)` is a simplex in the `\(VR(r)\)` simplicial complex iff `$$d(v_i,v_j)\leq r\quad\text{ for all }\quad i,j\in\{0,\ldots,n\}.$$` --- background-image: url("../../tda/figs/barcode_vr1.svg") background-size: 600px background-position: 50% 90% # Summarizing results in a barcode .pull-left[ <img src="../../tda/figs/vr1_summary.svg" width="400" style="display: block; margin: auto;" /> ] .pull-right[ - Five connected components are born from the get-go - Four of them merge into one at `\(r_4\)` - A hole is born with this merge - This hole eventually is filled with triangles at `\(r_6\)`. ] --- # Persistence diagrams .pull-left[ <img src="../../tda/figs/persistence_diagrams.svg" width="400" style="display: block; margin: auto;" /> ] .pull-right[ - Consider `\(C_{i,j;q}\)` all the `\(q\)`-classes that are born at time `\(i\)` and die at time `\(j\)` - The associated persistence diagram `\(\text{dgm}(\mathcal{F})\)` is a multiset `\(\text{dgm}(\mathcal{F}) = \bigcup_{q=0}^{n}\{(i,j) : \alpha_{i,j;q}\in C_{i,j;q}\}\cup\textrm{diag}\)` ] --- background-image: url("../../tda/figs/bottleneck_diagram.svg") background-size: 600px background-position: 50% 90% # Bottleneck distance and stability - Consider `\(\text{dgm}_1\)`, `\(\text{dgm}_2\)` two persistence diagrams. The *bottleneck distance$ between them is `$$d_B(\text{dgm}_1,\text{dgm}_2) = \inf_\gamma\sup_{p\in\text{dgm}_1}\|p-\gamma(p)\|_\infty.$$` - Where `\(\gamma\)` is a bijection between the `\(q\)`-th `\(\text{dgm}_1\)` and `\(\text{dgm}_2\)` - **Persistence diagrams are stable under the bottleneck distance!** --- ### Quantify the distribution of oil glands using Persistent Homology <div class="row"> <div class="column" style="max-width:30%"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L00_frontpic.jpg"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L00_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L01_frontpic.jpg"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L01_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L02_frontpic.jpg"> <img style="padding: 0 0 0 0;" src="../figs/M04_CRC3752_12B-28-7_L02_phlifetime.jpg"> </div> </div> Persistent Homology ( `\(H_0\)` and `\(H_1\)`) computed with Ripser in `scikit-tda`. --- ### Quantify the distribution of oil glands using Persistent Homology <div class="row"> <div class="column" style="max-width:30%"> <img src="../figs/M01_CRC3226_12B-27-1_L00_frontpic.jpg"> <img src="../figs/M01_CRC3226_12B-27-1_L00_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/M01_CRC3226_12B-27-1_L01_frontpic.jpg"> <img src="../figs/M01_CRC3226_12B-27-1_L01_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/M01_CRC3226_12B-27-1_L02_frontpic.jpg"> <img src="../figs/M01_CRC3226_12B-27-1_L02_phlifetime.jpg"> </div> </div> Persistent Homology ( `\(H_0\)` and `\(H_1\)`) computed with Ripser in `scikit-tda`. --- ### Quantify the distribution of oil glands using Persistent Homology <div class="row"> <div class="column" style="max-width:30%"> <img src="../figs/C02_CRC3919_18A-24-1_L00_frontpic.jpg"> <img src="../figs/C02_CRC3919_18A-24-1_L00_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/C02_CRC3919_18A-24-1_L01_frontpic.jpg"> <img src="../figs/C02_CRC3919_18A-24-1_L01_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/C02_CRC3919_18A-24-1_L02_frontpic.jpg"> <img src="../figs/C02_CRC3919_18A-24-1_L02_phlifetime.jpg"> </div> </div> Persistent Homology ( `\(H_0\)` and `\(H_1\)`) computed with Ripser in `scikit-tda`. --- ### Quantify the distribution of oil glands using Persistent Homology <div class="row"> <div class="column" style="max-width:30%"> <img src="../figs/WR02_CRC3877_12B-44-13_L00_frontpic.jpg"> <img src="../figs/WR02_CRC3877_12B-44-13_L00_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/WR02_CRC3877_12B-44-13_L01_frontpic.jpg"> <img src="../figs/WR02_CRC3877_12B-44-13_L01_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/WR02_CRC3877_12B-44-13_L02_frontpic.jpg"> <img src="../figs/WR02_CRC3877_12B-44-13_L02_phlifetime.jpg"> </div> </div> Persistent Homology ( `\(H_0\)` and `\(H_1\)`) computed with Ripser in `scikit-tda`. --- ### Quantify the distribution of oil glands using Persistent Homology <div class="row"> <div class="column" style="max-width:30%"> <img src="../figs/WR08_CRC3661_18B-16-5_L00_frontpic.jpg"> <img src="../figs/WR08_CRC3661_18B-16-5_L00_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/WR08_CRC3661_18B-16-5_L01_frontpic.jpg"> <img src="../figs/WR08_CRC3661_18B-16-5_L01_phlifetime.jpg"> </div> <div class="column" style="max-width:30%"> <img src="../figs/WR08_CRC3661_18B-16-5_L02_frontpic.jpg"> <img src="../figs/WR08_CRC3661_18B-16-5_L02_phlifetime.jpg"> </div> </div> Persistent Homology ( `\(H_0\)` and `\(H_1\)`) computed with Ripser in `scikit-tda`.