class: center, middle, inverse, title-slide .title[ # Characterizing spatial patterns and distributions ] .subtitle[ ## with Topological Data Analysis (TDA) ] .author[ ### <strong>Erik Amézquita</strong>, Sutton Tennant, Sandra Thibivillers, Sai Subhash<br>Benjamin Smith, Samik Bhattacharya, Jasper Kläver, Marc Libault<br>—<br>Division of Plant Science & Technology<br>Department of Mathematics<br>University of Missouri<br>— ] .date[ ### 2024-10-05 ] --- # Different **genes** in different **cells** follow different **spatial patterns** of expression .pull-left[  Some genes seem uniformly distributed ] .pull-right[  Other genes are more ring-like ] - Different cells have different sizes, shapes, and orientations --- # We use Topological Data Analysis (TDA)! 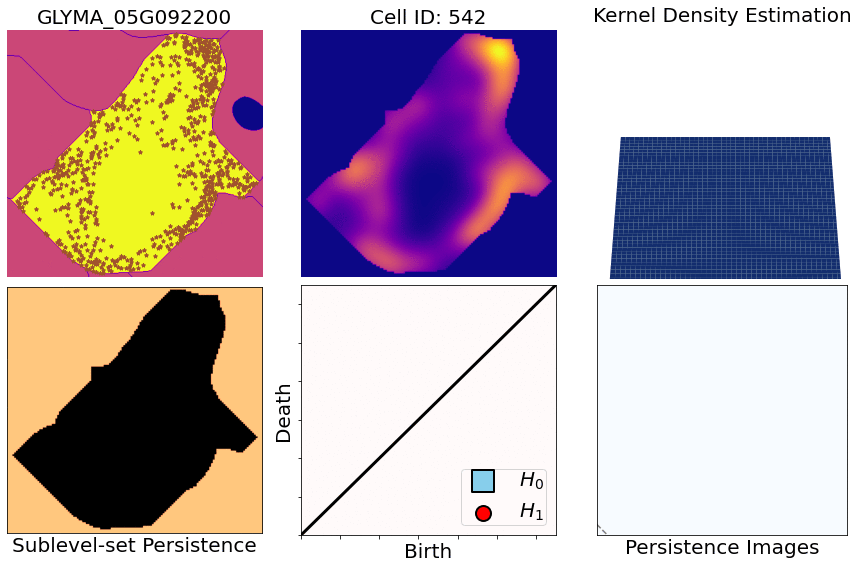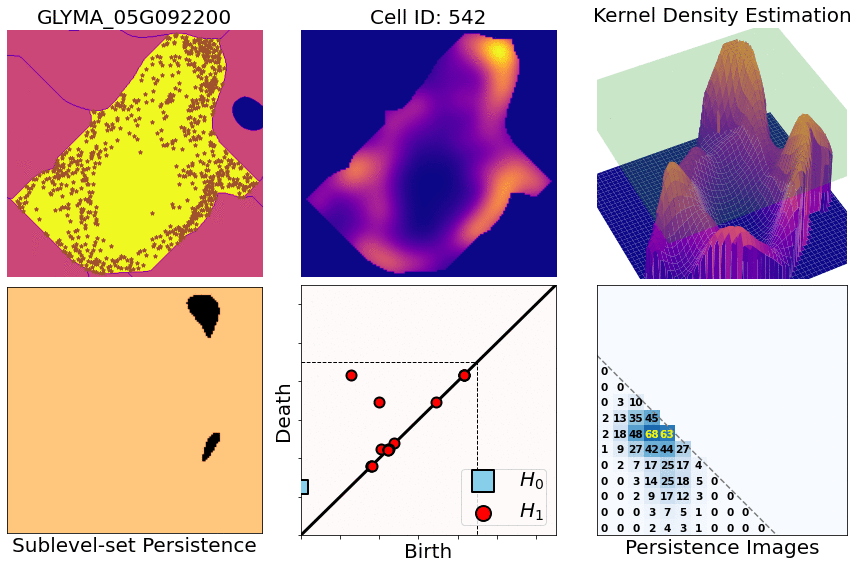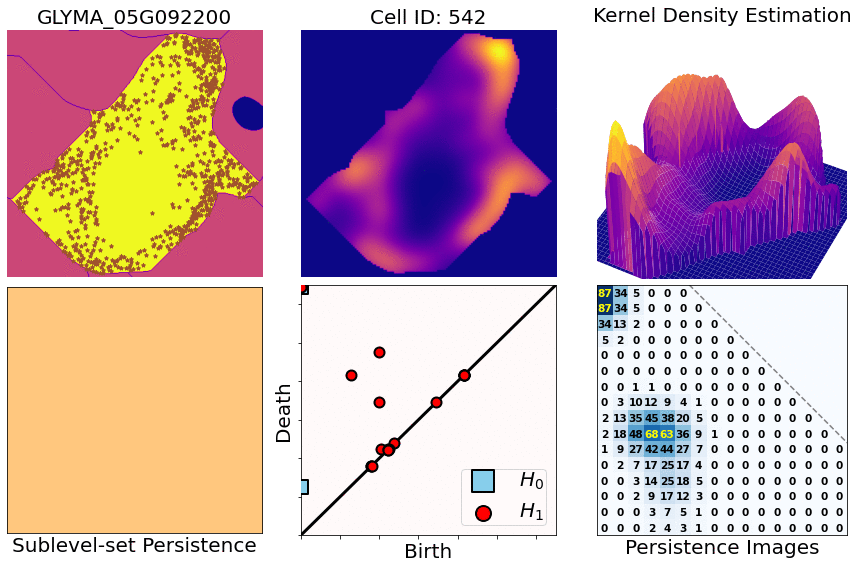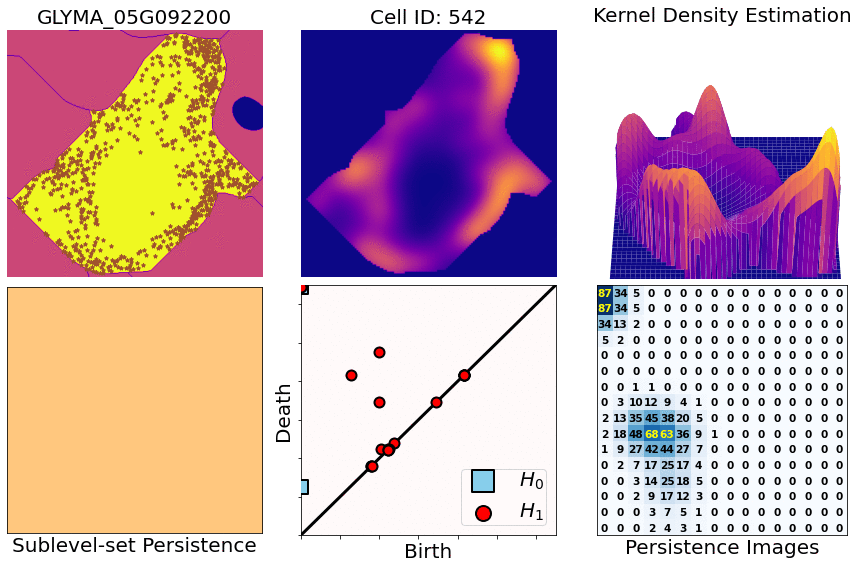 --- class: inverse, middle, center # [1] Molecular Cartography™ # & # Kernel Density Estimators (KDEs) [2] Quantify these heatmaps via Topological Data Analysis (TDA) [3] Link back to biological context --- # Molecular Cartography™  - Soybean nodule 10µm thick cross-sections. - (X,Y,Z) coordinates for 3.7M+ cytosolic transcripts. - 97 genes (including 10 bacterial ones) → 2 genes - 2938 cells → 918 infected ones. --- # Kernel Density Estimations (KDEs) 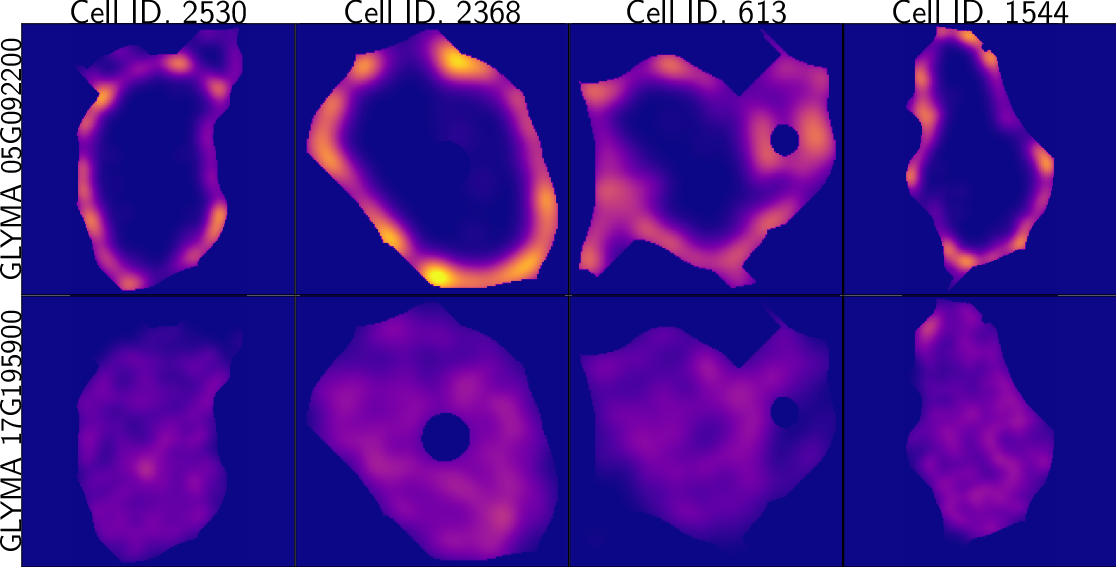 - **Normalize by cell AND gene:** The sum of all transcripts OF ALL genes add to 100% - Each gene adds to a certain percentage - The sum of all genes add to 100% - Compares absolute concentrations --- class: inverse, middle, center [1] Molecular Cartography™ & Kernel Density Estimations # [2] Quantify the shape of these heatmaps ## Topological Data Analysis (TDA) and persistent homology [3] Link back to biological context --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Sublevel filter → Persistence Diagrams  --- # Same idea for higher dimensions 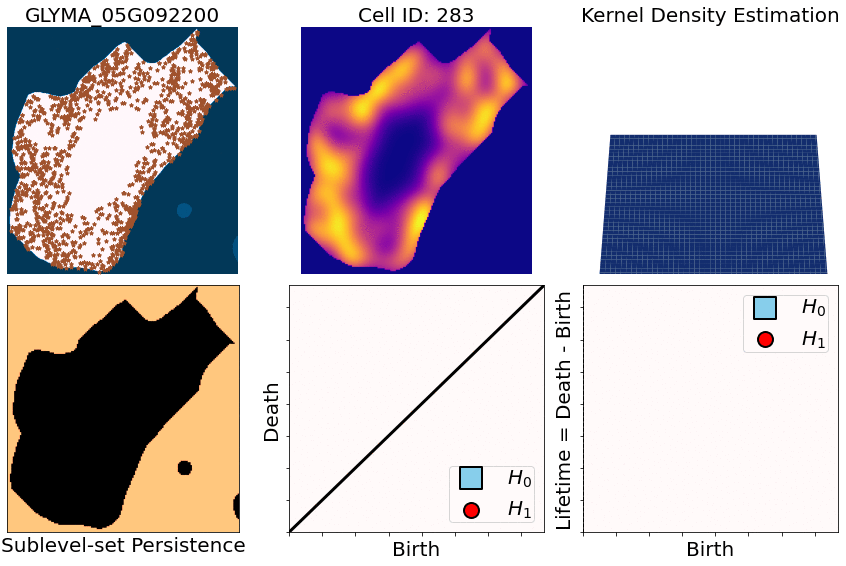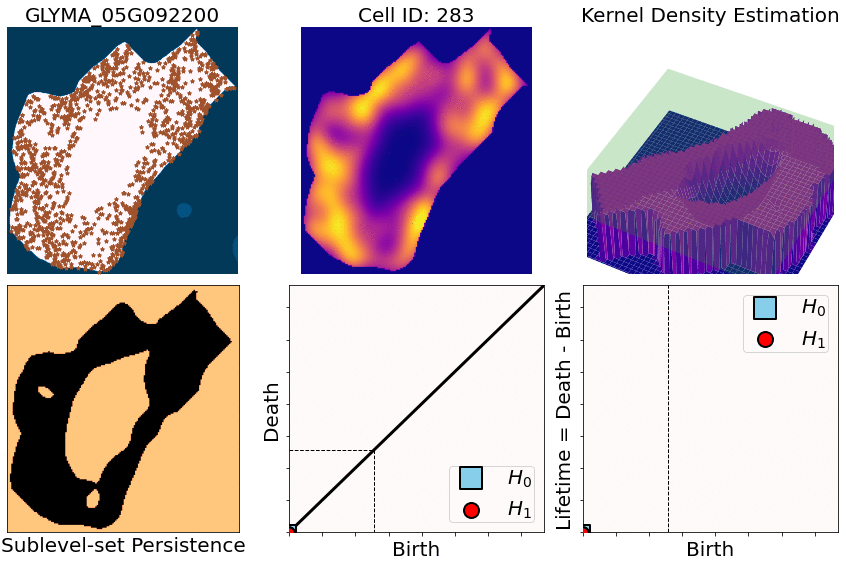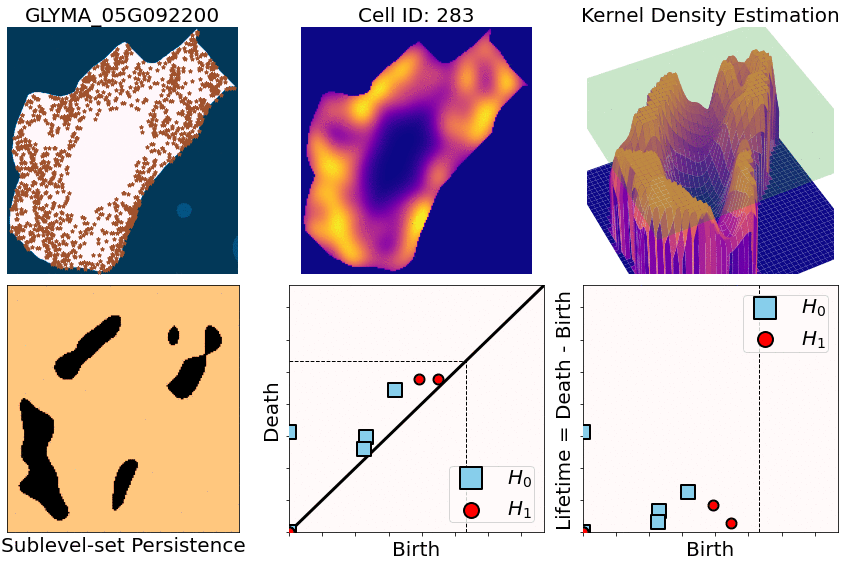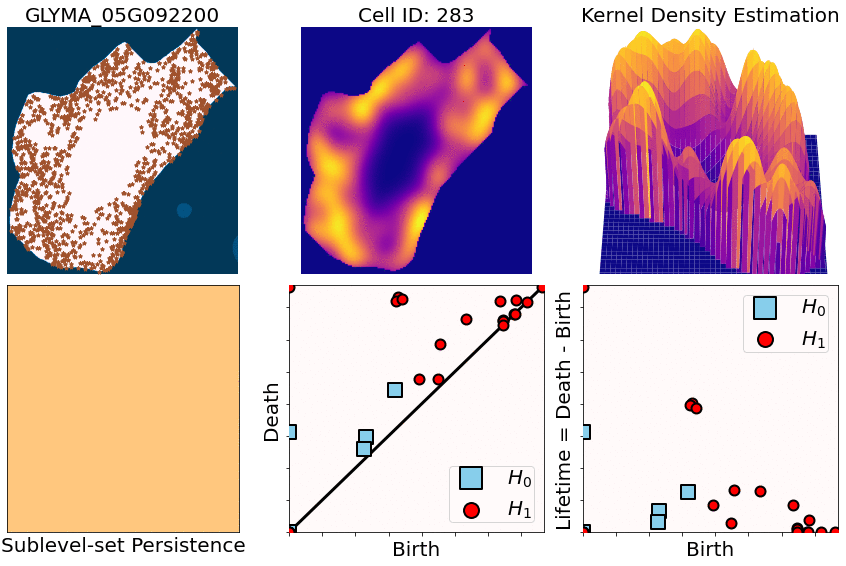 --- # Same idea for higher dimensions  --- # Rotate 45 degs for Persistence Images  --- # Rotate 45 degs for Persistence Images  --- # Mathematical justifications - **Definition:** Given two persistence diagrams `\(D_1, D_2\)`, for `\(1\leq p<\infty\)`, we define the *p-Wasserstein* distance between them as `$$W_p(D_1, D_2) := \inf_{\gamma:D_1\to D_2}\left(\sum_{u\in D_1} \left\| u-\gamma(u) \right\|_\infty^p\right)^{1/p},$$` where the infimum is over all possible bijections `\(\gamma: D_1\to D_2\)`. - **Theorem [[Mileyko *et al* (2011)](https://doi.org/10.1088/0266-5611/27/12/124007)]:** For nice filtrations, the persistence diagrams are Wasserstein-stable under small perturbations of the data they summarize. - **Theorem [[Adams *et al.* (2017)](http://jmlr.org/papers/v18/16-337.html)]:** The persistence image `\(I(D)\)` of a persistence diagram `\(D\)` with Gaussian distributions is stable with respect to the 1-Wasserstein distance between diagrams. ### If the overall shape/pattern is perturbed a little bit, then the resulting persistence images are perturbed only a little bit as well --- class: inverse, middle, center [1] Molecular Cartography™ & Kernel Density Estimations [2] Quantify the shape of these heatmaps # [3] Link back to biological context ## Never underestimate the power of PCA --- # Do TDA for all cell-gene combinations <img src="../figs/molecular_cartography_2x4.png" width="500" style="display: block; margin: auto;" /> <img src="../figs/persistence_images_2x4.png" width="600" style="display: block; margin: auto;" /> --- background-image: url("../figs/bw20_both_scale16_-_PI_1_1_1_pca_H1+2_cell_sample.png") background-size: 620px background-position: 75% 99% # PCA on all topological descriptors <img src="../figs/bw20_both_scale16_-_PI_1_1_1_pca_H1+2_gridded.png" width="250" style="display: block; margin: auto auto auto 0;" /> --- background-image: url("../figs/bw20_both_scale16_-_PI_1_1_1_pca_H1+2_kde_sample.png") background-size: 620px background-position: 75% 99% # Show me <img src="../figs/bw20_both_scale16_-_PI_1_1_1_pca_H1+2_gridded.png" width="250" style="display: block; margin: auto auto auto 0;" /> --- # Relate back to the biological context  - Senescent cells exhibit a distinct transcriptomic spatial pattern compared to the rest of population. - Loss of mRNA localization may be a lesser known contributor to cell senescence. --- # Discussion - Topological Data Analysis offers a robust way to encode the *shape of patterns* - Robust to differences in scale, underlying boundaries, or orientation - As long as you have a heatmap, you're good to go: - Even if you don't, you can make one via KDEs from individual spatial coordinates - Climate patterns? - Canopy patterns? - Cover crop patterns? - Species spatial distribution?  --- # Software used .pull-left[ - All the work has been done in python with mostly standard libries (`numpy`, `scikit-learn`, `matplotlib`, `pandas`, etc.) - KDEs computed efficiently with [`KDEpy`](https://kdepy.readthedocs.io/en/latest/) - Sublevel set filtration of images and persistence diagrams done with [`gudhi`](https://gudhi.inria.fr/)  ] .pull-right[ - Persistence Images computed with [`persim`](https://persim.scikit-tda.org/en/latest/)  ] --- class: inverse # Thank you! <div class="row"> <div class="column" style="max-width:60%; font-size: 15px;"> <img style="padding: 0 0 0 0;" src="https://bondlsc.missouri.edu/wp-content/uploads/2023/11/libault-returns-to-bond-lsc-as-plant-scientist-with-single-cell-focus-1.jpg"> </div> <div class="column" style="max-width:40%; font-size: 24px; line-height:1.25"> <p style="text-align: center;"><strong>Contact and slides:</strong></p> <p style="text-align: center;color:Blue">eah4d@missouri.edu</p> <p style="text-align: center;color:Blue">ejamezquita.github.io</p> <p style="font-size: 18px">Registration and travel support for this presentation was provided by the National Science Foundation and NSF Project 00087298.</p> </div> </div> <div class="row"> <div class="column" style="max-width:35%; font-size: 20px;"> <p style="font-size: 25px; text-align: center;"> Libault Lab (MU) </p> <ul> <li><strong>Marc Libault</strong></li> <li><strong>Sandra Thibivillers</strong></li> <li>Hengping Xu</li> <li>Sahand Amini</li> <li>Hong Fu</li> <li><strong>Sutton Tennant</strong></li> <li>Md Sabbir Hossain</li> <ul> </div> <div class="column" style="max-width:65%; font-size: 20px;"> <p style="font-size: 25px; text-align: center;"> With help from:</p> <ul> <li>Sai Subash (Nebraska-Lincoln)</li> <li>Benjamin Smith (UC Berkeley)</li> <li>Samik Bhattacharya (Resolve Biosciences)</li> <li>Jasper Kläver (Resolve Biosciences)</li> <ul> </div> </div>