class: center, middle, inverse, title-slide .title[ # Going between math, data science, plant biology, and education ] .subtitle[ ## My interdisciplinary journey ] .author[ ### Erik Amézquita<br>—<br>Division of Plant Science & Technology<br>Department of Mathematics<br>University of Missouri<br>— ] .date[ ### 2025-08-18 ] --- background-image: url("../../demat/figs/fam9_3.png") background-size: 100px background-position: 98% 2% # About me: From MX to MI to MO at MU ## I work across multiple disciplines and countries .left-column[  ] .right-column[ - 2013 - 2018 : Licenciatura (Bachelor): Mathematics @ Universidad de Guanajuato and CIMAT. Thesis focused on Topological Data Analysis applied to archaeology. - 2018 - 2023 : PhD: CMSE @ MSU. Dissertation: Exploring the mathematical shape of plants. - 2023 - 2025 : ~~PFFFD~~ ~~PFFIE~~ PFF Postdoctoral Fellow: Plant Science (80%) / Mathematics (20%) @ University of Missouri (MU). - 2025 - present: Assistant Professor: Plant Science (100%) / Math (0%) @ MU. ] --- background-image: url("../../barley/figs/seed.png") background-size: 325px background-position: 99% 99% class: middle # Roadmap for today 1. My research before CMSE - Mixing TDA (Topological Data Analysis) with archaeology 1. My research while at CMSE - Mixing TDA with plant biology 1. My research right after CMSE - Jack of all trades, master of none 1. Current projects that excite me - Taking a leaf from CMSE 201 --- background-image: url("../../img/cmse_logo.svg") background-size: 300px background-position: 50% 45% class: inverse, center, middle # A mathematician by training, a data scientist by trade, and a plant biologist by ~~collaboration~~ osmosis <p style="opacity:0">a<br>aa<br>aa<br>aa<br>aa<br>aa<br>a</p> <img src="https://png.pngtree.com/png-vector/20220617/ourmid/pngtree-diagram-showing-osmosis-in-plant-cell-natural-education-plants-vector-png-image_37088024.png" width="400" style="display: block; margin: auto;" /> --- background-image: url("../../demat/figs/ecg_results.png") background-size: 700px background-position: 50% 80% # 2017: Pre-CMSE → TDA ⊕ Archaeology Topological Data Analysis for pre-Columbian mask classification -- .pull-left[ <img src="../../demat/figs/barrett_poster-1.jpg" width="310" style="display: block; margin: auto;" /> ] -- .pull-right[  ``` r *- Lessons learned for better * future interdisciplinary * collaborations *- Always include the other * discipline experts at * every stage of the analysis ``` ] --- background-image: url("../../arabidopsis/figs/PFig1.png") background-size: 900px background-position: 50% 70% # 2018: CMSE → TDA ⊕ Plant Biology Mathematically quantify the shape of plants with Liz Munch and Dan Chitwood -- .pull-left[  <img src="../../arabidopsis/figs/DSCN0077.jpg" width="500" style="display: block; margin: auto;" /> ] .pull-right[ 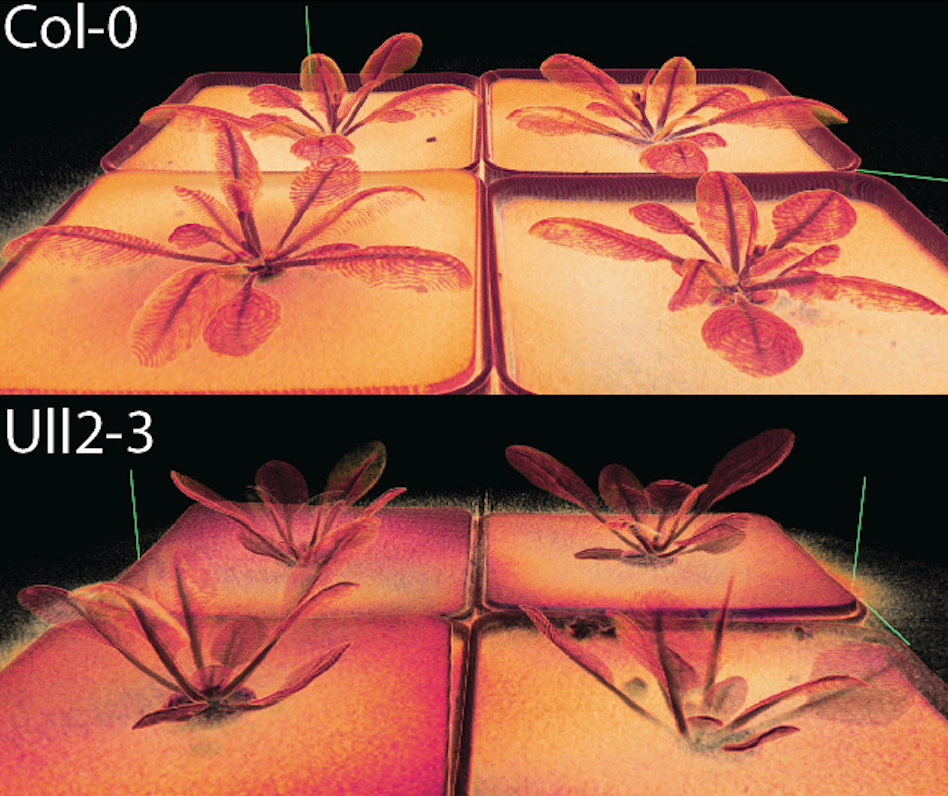 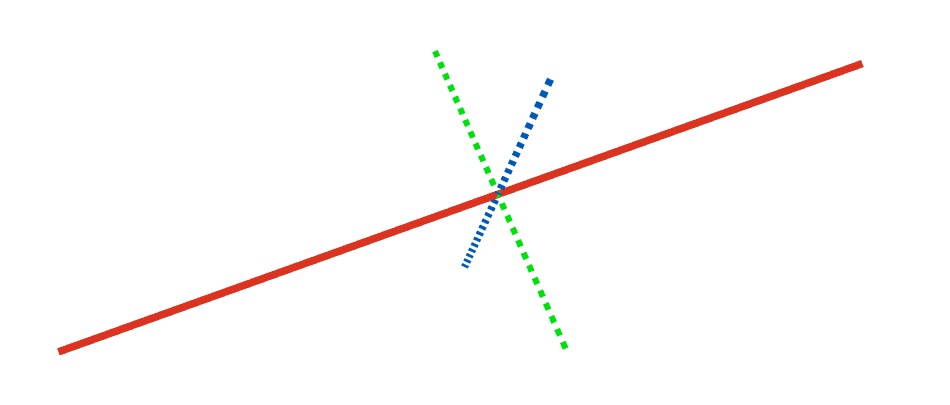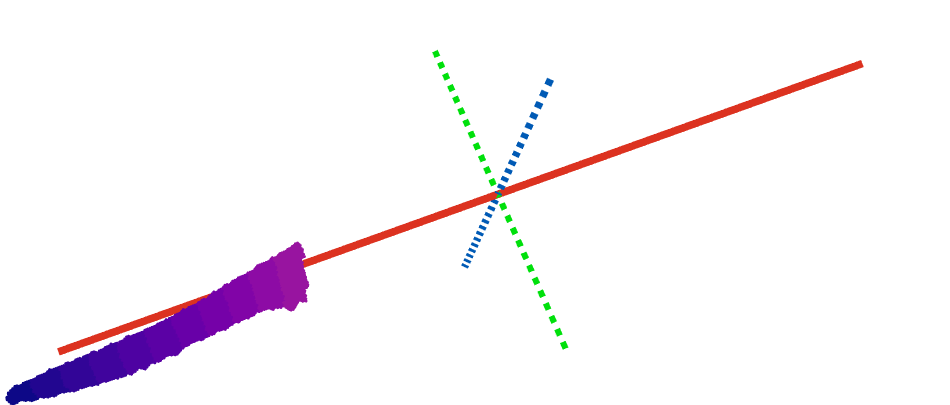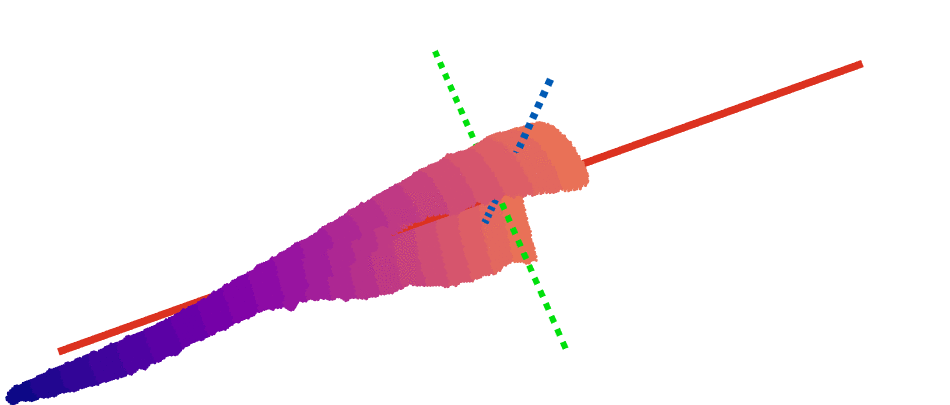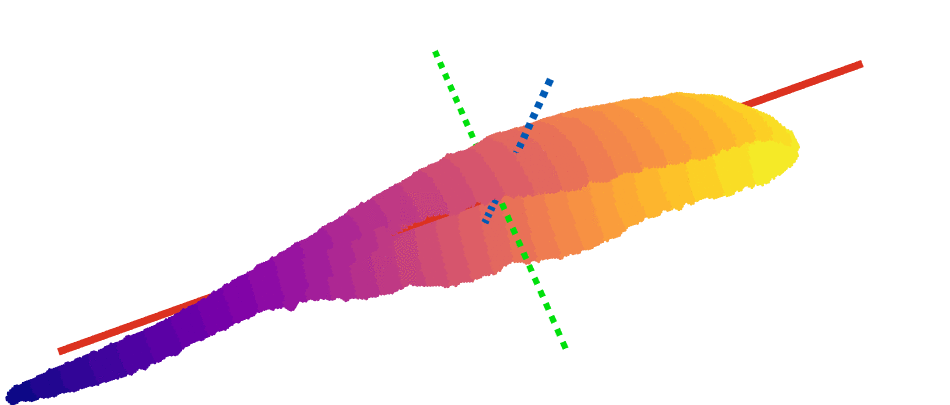 ] --- # Come for the math, stay for the plants <div class="row" style="font-family: 'Yanone Kaffeesatz'; font-size:22px;"> <div class="column" style="max-width:33%"> <p style="line-height:0;text-align: center; font-size:28px">The shape of adaptability</p> <img style="padding: 0 70px;" src="../../barley/figs/S017_L3_1.gif"></img> <img style="padding: 0 55px;" src="../../barley/figs/ecc_X.gif"></img> <p style="text-align: center;">Topological Data Analysis</p> <p style="text-align: center;">Euler Characteristic Transform</p> </div> <div class="column" style="max-width:33%"> <p style="line-height:0;font-size:28px;text-align: center;">The shape of development</p> <img style="padding: 0 60px;" src="../../citrus/figs/SR01_L01_black_exocarp.gif"></img> <img style="padding: 0 27px;" src="../../citrus/figs/SW03_CRC1241-B_12B-4-3_L00_lambproj_v.jpg"></img> <p style="text-align: center;">Ellipsoidal modeling</p> <p style="text-align: center;">Directional statistics</p> </div> <div class="column" style="max-width:33%"> <p style="line-height:0;font-size:28px;text-align: center;">The shape of domestication</p> <img style="padding: 0 35px;" src="../../walnuts/figs/2014SBa_R5_T81_meat_2.gif"></img> <img style="padding: 0 10px;" src="../../walnuts/figs/kernel_volume_vs_nut_volume.png"></img> <p style="text-align: center;">Allometry of multiple tissues</p> <p style="text-align: center;">Convexity indices</p> </div> </div> --- class: inverse, middle, center # 2023: Post-CMSE → Math ⊕ Plants ## It is far easier to switch countries than to switch disciplines ## But CMSE trained me for the cultural shock <img src="../../img/homotopy_botany.png" width="500" style="display: block; margin: auto;" /> <p style="font-size: 10px; text-align: center; color: Grey;">Credits: <a href="http://dx.doi.org/10.2140/agt.2005.5.237">Kaufmann (2005)</a></p> --- # Tracking how a vegetarian plant wiggles <video width="900" controls> <source src="../../cuscuta/video/9am_Inc_Rep_3_redone.mp4" type="video/mp4"> </video> - *Cuscuta campestris* inoculated at different times of the day for 24h - In collaboration with Soyon Park --- ## Phenotyping movement: how *wiggly* is a wiggle? <video width="900" controls> <source src="../../cuscuta/video/4pm_rep7_plant_01.mp4" type="video/mp4"> </video> - Mathematically model how *Cuscuta* moves under various environmental conditions to ultimately stop it from attaching to crops in the first place. --- ## Phenotyping patterns: How *patterny* is a pattern?  - Beyond gene expression counts: Spatial segregation and asymmetrical distribution of mRNA across the cytosol in the soybean nodule. - Molecular Cartography™ data provided by the Libault Lab --- ## Topological Data Analysis to quantify our intuition  --- background-image: url("../../mcarto/figs/scale32_-_PI_1_1_1_H1+2_synthetic_30_clusters.jpg") background-size: 900px background-position: 50% 80% ## Define a morphospace of transcriptomic patterns -- <p style="opacity:0">a<br>aa<br>aa<br>aa<br>aa<br>aa<br>aaa<br>aa<br>a</p> <img src="../../mcarto/figs/scale32_-_PI_1_1_1_H1+2_synthetic_pca_30_clusters.jpg" width="600" style="display: block; margin: auto;" /> --- background-image: url("../../tutorials/figs/mizzou_math_drp.png") background-size: 250px background-position: 99% 1% ## More data, more shapes, more phenotyping .pull-left[ Academic social network analysis of the IPG (Interdisciplinary Plant Group) <img src="../../tutorials/figs/knehansIPG.jpg" width="300" style="display: block; margin: auto;" /> Similar work to be done with gene coexpression and biota interaction networks. ] .pull-right[ Use TDA to analyze geographical patterns across the state. <p style="font-size: 10px; text-align: center; color: Grey;">Credits: <a href="https://doi.org/10.1137/19M1241519">Feng and Porter (2021)</a></p> <img src="../../tda/figs/025-imperial.png" width="180" style="display: block; margin: auto;" /><img src="https://www.researchgate.net/publication/362833654/figure/fig2/AS:11431281098979080@1669173266225/Two-persistence-diagrams-for-the-simulation-shown-in-Fig-1-The-blue-crosses-represent.png" width="180" style="display: block; margin: auto;" /><img src="../../psd/figs/pavement_plasma.jpg" width="180" style="display: block; margin: auto;" /> A voting district looks like a plant cell. ] --- background-image: url("https://plantsandpython.github.io/PlantsAndPython/_images/plants_python_logo.jpg") background-size: 180px background-position: 99% 1% class: inverse, center, middle # Shaping the next generation of interdisciplinary scientists ## Large amounts of data require large amounts of people ## PLNT_SCI 2500: Data Science for Life Sciences I  --- ## Taking a leaf from CMSE 201 <p align="center"> <iframe width="800" height="550" src="../../tutorials/plnt_2500/Day-13_In-Class_Regression-INSTRUCTOR.html" title="Day10"> </iframe> </p> --- ## PLNT_SCI 2500: With data from Missouri faculty! <p align="center"> <iframe width="800" height="550" src="../../tutorials/plnt_2500/Day-20_In-Class_AdvancedPlotting-INSTRUCTOR.html" title="Day10"> </iframe> </p> --- # The shape of things to come .left-column[   ] .right-column[ - Make the teaching of data science in life sciences a scientific endeavor in itself. - More **translations** of Topological Data Analysis and other mathematical techniques to quantify plant biology-inspired data - Explore more applications of complex network theory to biology and meta-biology contexts. **Adverstisement** - I will be looking for PhD students to start next year working with me at the intersection of math, data science, and plant science ] <img src="https://gradschool.missouri.edu/wp-content/uploads/2019/02/MU_UnitSig_GraduateSchool_rgb_std_horiz.png" width="500" style="display: block; margin: auto;" /> --- background-image: url("https://upload.wikimedia.org/wikipedia/commons/4/4a/University_of_Missouri_logo.svg") background-size: 60px background-position: 99% 1% class: inverse ## Thank you! .pull-left[ **Research while at CMSE** - Liz Munch - Dan Chitwood - Michelle Quigley - Tim Ophelders - Jacob Landis - Dan Koenig - Danelle Seymour - Pat Brown **Cuscuta rhythm and locomotion** - Max Bentelspacher - Supral Adhikari - Jaime Barros-Rios - Joseph Lynch - So-Yon Park **mRNA sub-cellular localization** - Sutton Tennant - Sandra Thibivillers - Sai Subhash - Benjamin Smith - Samik Bhattacharya - Jasper Kläver - Marc Libault ] .pull-right[ **Collaboration of the IPG network** - Ethan Lenhardt - Sophia Knehans - Roberto Herrera - David Braun **Data Science for Life Sciences I** - Kent Shannon - Andrew Scaboo - Jianfeng Zhou - Debbie Finke **Other ongoing projects** - Leyre Urmeneta - Laura Martins - Mather Khan - David Mendoza-Cozatl - Jie Zhu **More details** <p style="font-size: 20px; text-align: center; color: Blue;">ejamezquita.github.io/</p> <p style="font-size: 20px; text-align: center; color: Blue;">eah4d@missouri.edu</p> ]