Characterizing trascriptomic spatial patterns
Different genes in different cells follow different spatial patterns of expression. We can clearly tell by eye. Our gut feeling tells us that some patterns look rather uniform, others spotty, and others pretty circular.
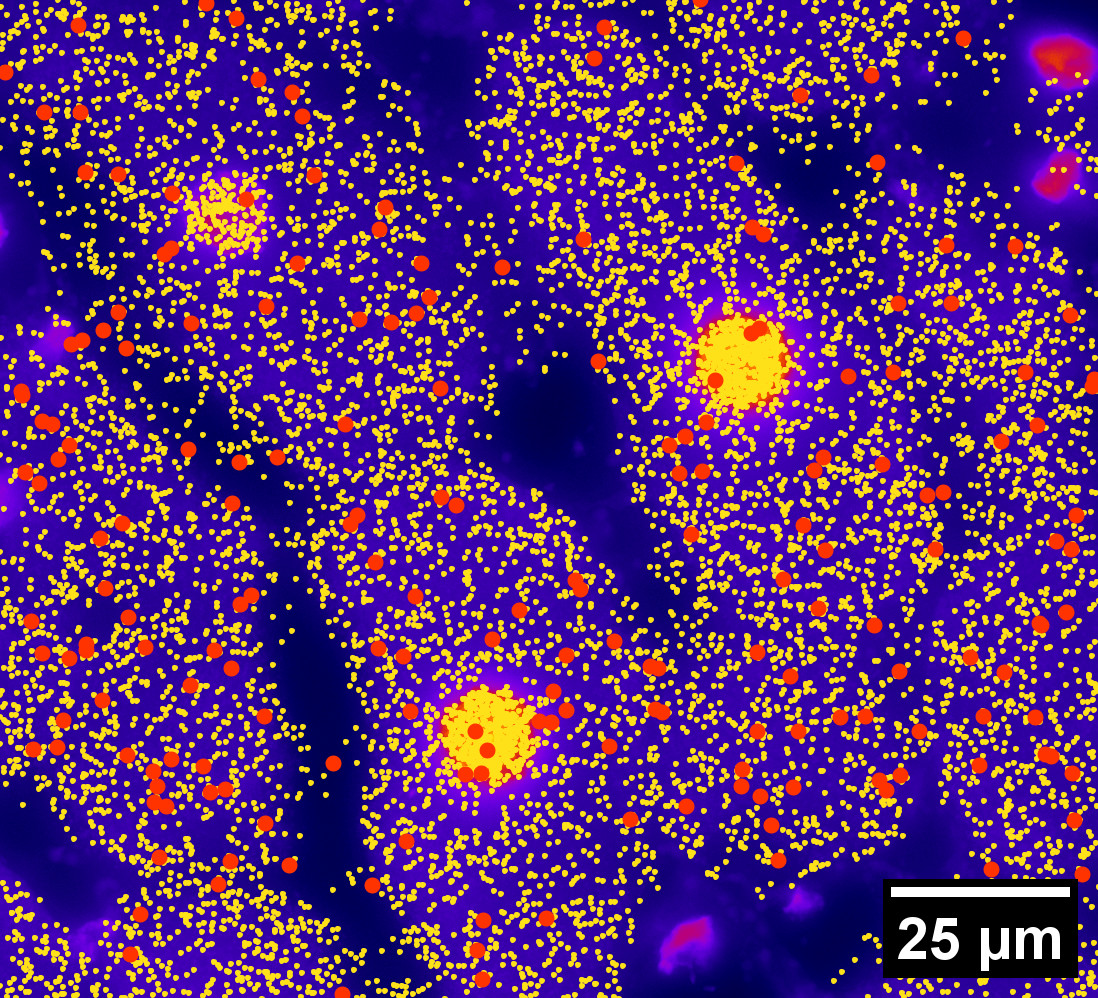
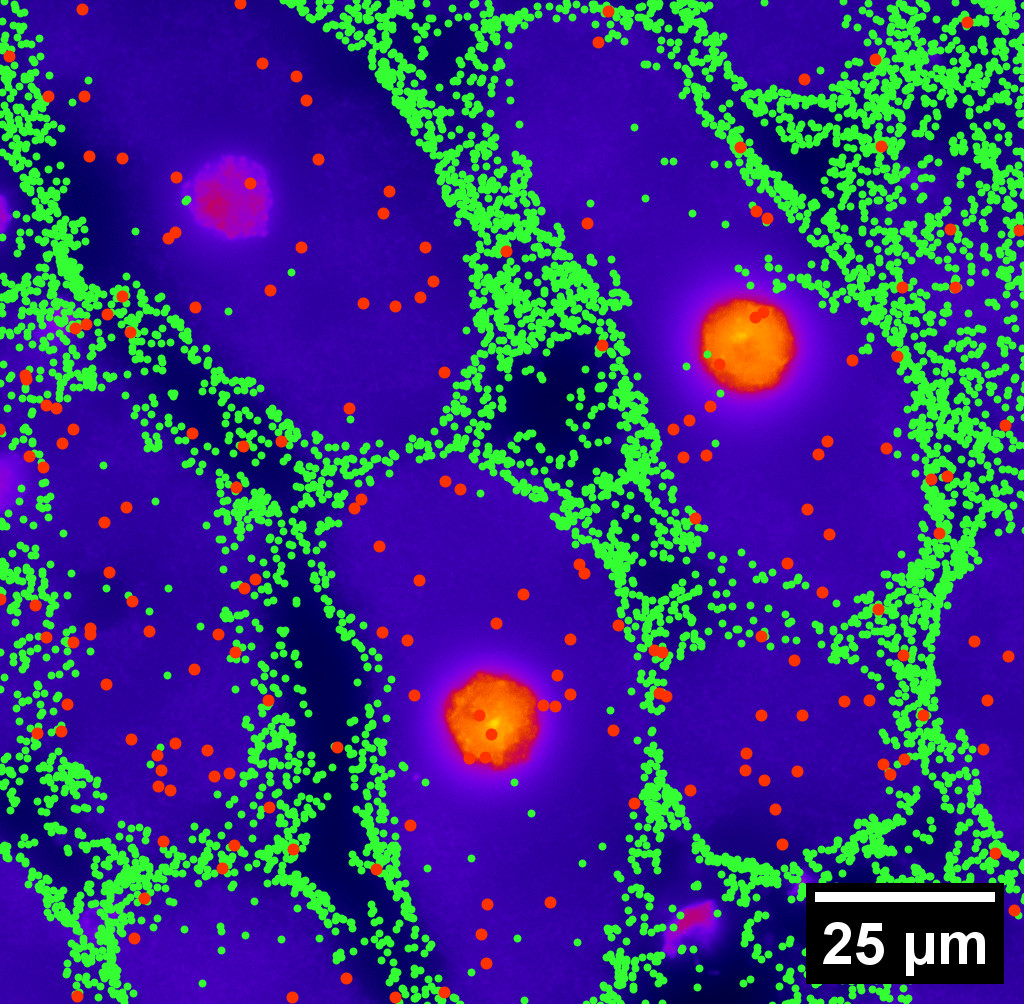
Understanding cell-gene spatial patterns will further our biological knowledge. But how do we quantify these patterns?
We need a platform that is robust enough to take into account all possible cell shapes, sizes, and orientations. A pipeline that can work regardless of the amount of transcripts. We turn thus to Topological Data Analysis (TDA).
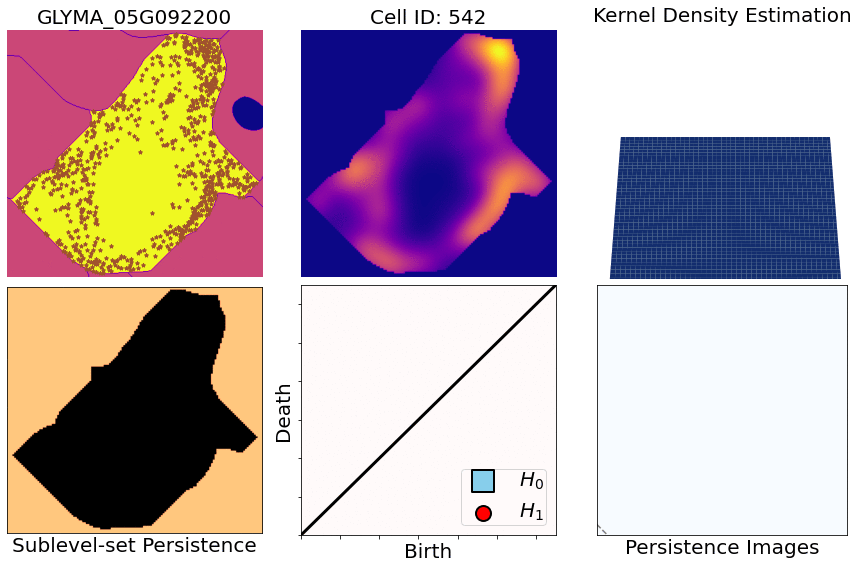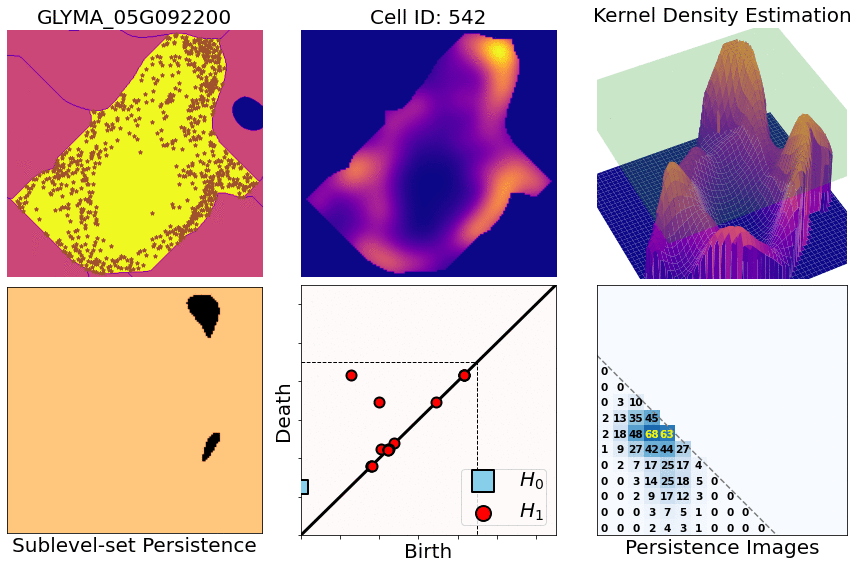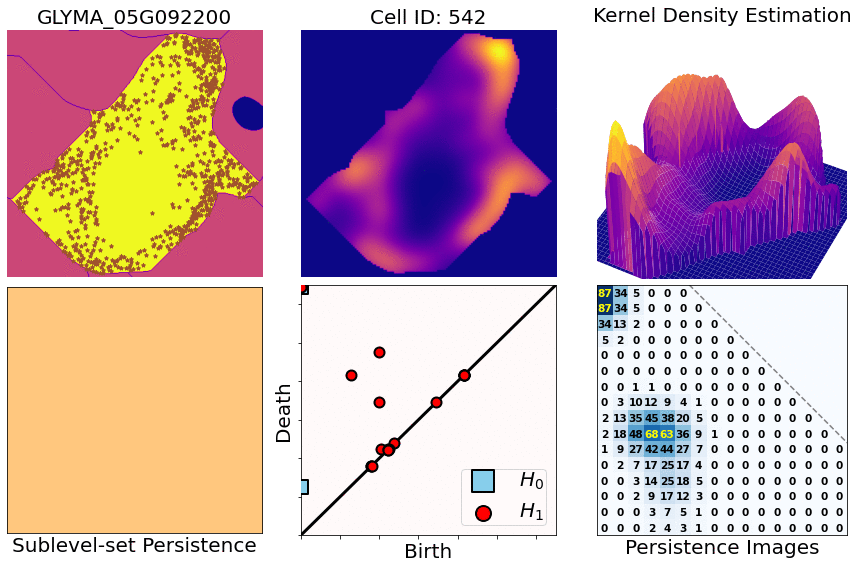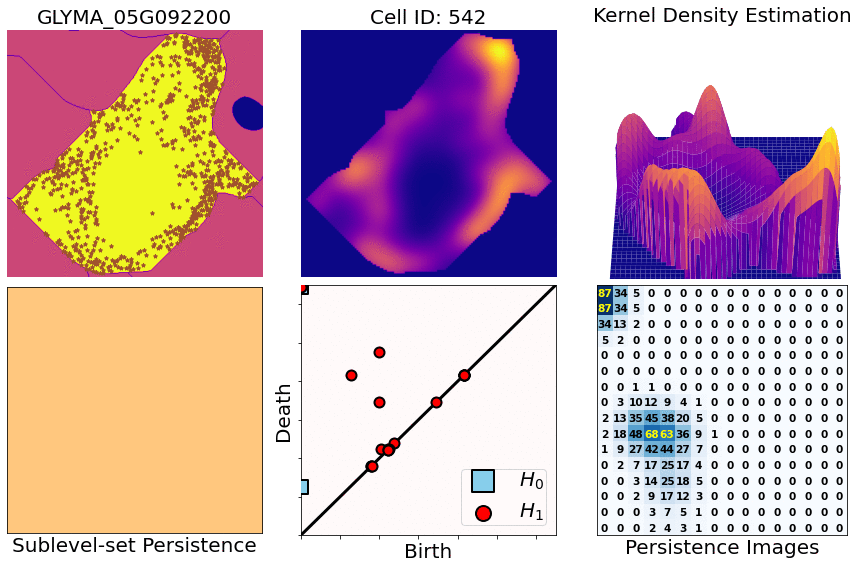
- You start with X,Y,Z coordinates of transcripts for one specific gene and one specific cell
- Compute heatmaps via KDEs (kernel density estimates)
- Compute persistence diagrams via sublevel set persistence (for both 1- and 2-dimensional holes [ H1 and H2 respectively])
- Transform those diagrams into persistence images (for both 1- and 2-dimensional diagrams)
- Concatenate both dimensions and flatten them into a single row. Then attach all the rows for all the cells and genes: this produces a large matrix (4800 rows [cell/gene combinations] x 300 columns [topological features], approx)
- Compute the PCA of this matrix. Between the first two PCs we are able to capture more than 75% of the total topological variance when it comes to the spatial distributions of transcripts (or the shape of the heatmaps)
(One day of this I will write a better summary of this topic here.)
¡Preprint: In progress!
—
Code Available: Github repository with a series of Python-based Jupyter notebooks detailing the image-processing pipeline we wrote to detect Cuscuta from skewer and background. It also contains code on how time series were created and later analyzed.
—As slides: Presented at AATRN. Applied Algebraic Topology Research Network Seminar. Virtual. May 2025.
This is the recording of my talk.
As a poster: Presented at the CAFNR Research Symposium. University of Missouri. Columbia, Missouri. October 2024.
——————————
Other research projects
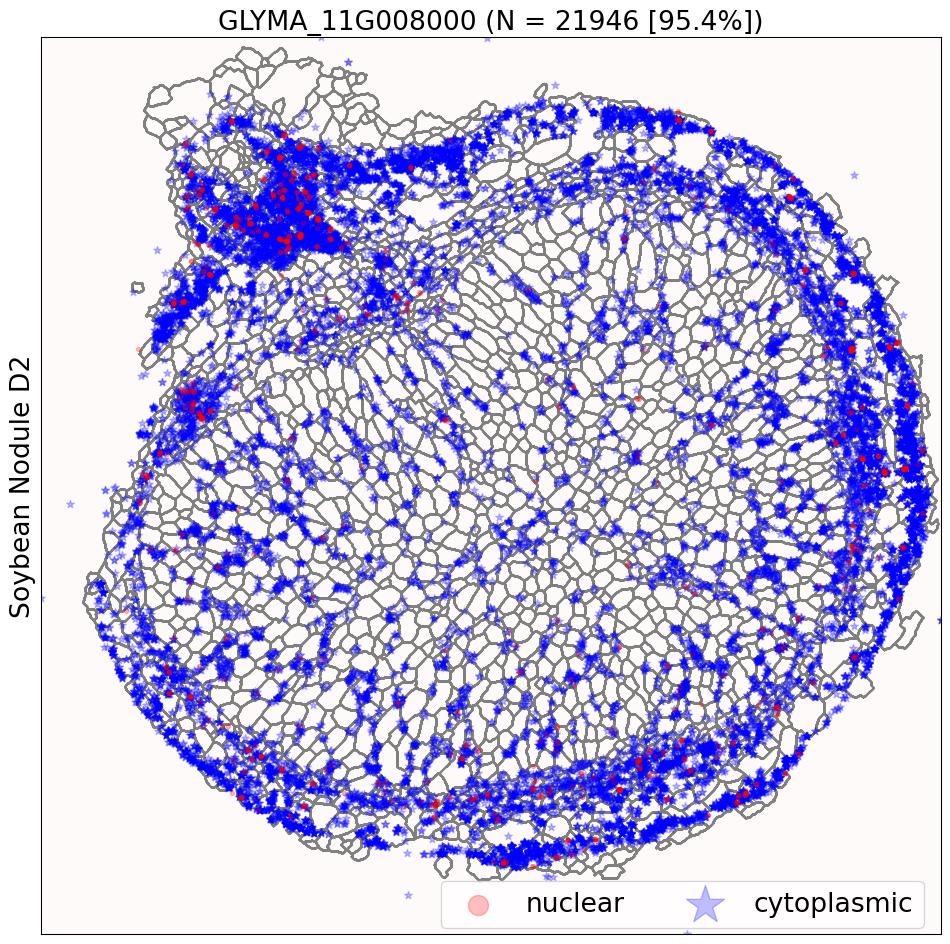
- Sub-cellular transcriptomic patterns
- The early dodder gets the host
- The crackability of walnuts: all about shape, in a nutshell
- Quantification of barley grain morphology
- Global disparities in plant biology research
- Mapper to unravel the shape of omics data
- The intersection of Topological Data Analysis and Biology
- The shape of citrus fruits and modeling their oil gland distribution
- Archaeological artifact classification and the Euler Characteristic